Difference between revisions of "PurB"
| Line 82: | Line 82: | ||
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P12047 P12047] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P12047 P12047] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU06440] |
* '''E.C. number:''' [http://www.expasy.org/enzyme/4.3.2.2 4.3.2.2] | * '''E.C. number:''' [http://www.expasy.org/enzyme/4.3.2.2 4.3.2.2] | ||
Revision as of 03:57, 25 June 2009
- Description: adenylsuccinate lyase
| Gene name | purB |
| Synonyms | |
| Essential | no |
| Product | adenylsuccinate lyase |
| Function | purine biosynthesis |
| Metabolic function and regulation of this protein in SubtiPathways: Purine salvage, Purine synthesis, Nucleotides (regulation), Stress | |
| MW, pI | 49 kDa, 5.822 |
| Gene length, protein length | 1293 bp, 431 aa |
| Immediate neighbours | purK, purC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 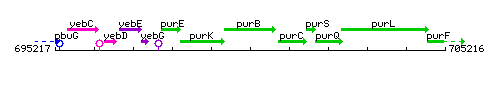
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU06440
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: N(6)-(1,2-dicarboxyethyl)AMP = fumarate + AMP (according to Swiss-Prot)
- Protein family: Adenylosuccinate lyase subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure: 1F1O
- Swiss prot entry: P12047
- KEGG entry: [3]
- E.C. number: 4.3.2.2
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Lars Engholm Johansen, Per Nygaard, Catharina Lassen, Yvonne Agersø, Hans H Saxild
Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL).
J Bacteriol: 2003, 185(17);5200-9
[PubMed:12923093]
[WorldCat.org]
[DOI]
(P p)
M Weng, P L Nagy, H Zalkin
Identification of the Bacillus subtilis pur operon repressor.
Proc Natl Acad Sci U S A: 1995, 92(16);7455-9
[PubMed:7638212]
[WorldCat.org]
[DOI]
(P p)
H H Saxild, P Nygaard
Regulation of levels of purine biosynthetic enzymes in Bacillus subtilis: effects of changing purine nucleotide pools.
J Gen Microbiol: 1991, 137(10);2387-94
[PubMed:1722815]
[WorldCat.org]
[DOI]
(P p)
D J Ebbole, H Zalkin
Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis.
J Biol Chem: 1987, 262(17);8274-87
[PubMed:3036807]
[WorldCat.org]
(P p)