Difference between revisions of "PucG"
| Line 26: | Line 26: | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yurG_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yurG_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=pucG_3341166_3342416_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:pucG_expression.png|500px]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | |||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
Revision as of 08:38, 23 April 2012
- Description: (S)-ureidoglycine-glyoxylate aminotransferase
| Gene name | pucG |
| Synonyms | yurG |
| Essential | no |
| Product | (S)-ureidoglycine-glyoxylate aminotransferase |
| Function | purine utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Purine catabolism, Nucleotides (regulation) | |
| MW, pI | 45 kDa, 5.42 |
| Gene length, protein length | 1248 bp, 416 aa |
| Immediate neighbours | pucA, pucF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 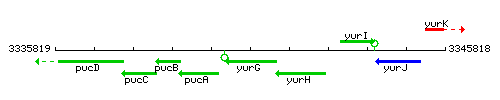
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed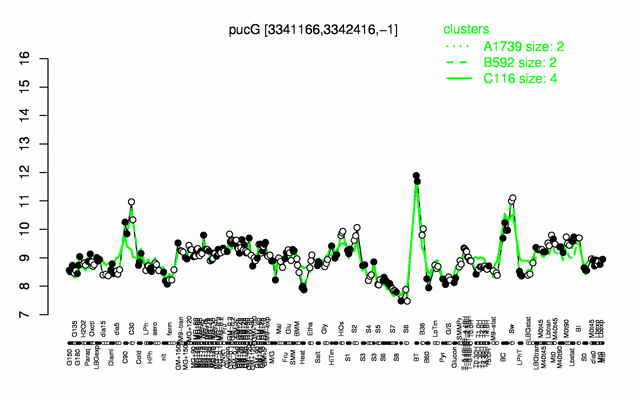
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU32520
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transamination between an unstable intermediate ((S)-ureidoglycine) and the end product of purine catabolism (glyoxylate) to yield oxalurate and glycine PubMed
- Protein family: class-V pyridoxal-phosphate-dependent aminotransferase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: O32148
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ileana Ramazzina, Roberto Costa, Laura Cendron, Rodolfo Berni, Alessio Peracchi, Giuseppe Zanotti, Riccardo Percudani
An aminotransferase branch point connects purine catabolism to amino acid recycling.
Nat Chem Biol: 2010, 6(11);801-6
[PubMed:20852637]
[WorldCat.org]
[DOI]
(I p)
Lars Beier, Per Nygaard, Hanne Jarmer, Hans H Saxild
Transcription analysis of the Bacillus subtilis PucR regulon and identification of a cis-acting sequence required for PucR-regulated expression of genes involved in purine catabolism.
J Bacteriol: 2002, 184(12);3232-41
[PubMed:12029039]
[WorldCat.org]
[DOI]
(P p)
A C Schultz, P Nygaard, H H Saxild
Functional analysis of 14 genes that constitute the purine catabolic pathway in Bacillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator.
J Bacteriol: 2001, 183(11);3293-302
[PubMed:11344136]
[WorldCat.org]
[DOI]
(P p)