Difference between revisions of "PtsI"
| Line 137: | Line 137: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' available in [[Stülke]] lab | + | * '''Mutant:''' available in [[Jörg Stülke]]'s lab: |
** GP864 (ermC) | ** GP864 (ermC) | ||
| − | ** GP778 (replacement of ''[[glcT]]'' and the ''[[ptsG]]-[[ptsH]]-[[ptsI]]'' operon by a spc cassette) | + | ** GP778 (replacement of ''[[glcT]]'' and the ''[[ptsG]]-[[ptsH]]-[[ptsI]]'' operon by a spc cassette), {{PubMed|22722928}} |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| − | ** pAG3 (His-tag), available in [[Galinier]] lab | + | ** pAG3 (His-tag) {{PubMed|9237995}}, available in [[Galinier]] lab |
| − | ** for expression, purification in ''E. coli'' (His-tag), in [[pWH844]]: pGP813 available in [[Stülke]] lab | + | ** for expression, purification in ''E. coli'' (His-tag), in [[pWH844]]: pGP813 available in [[Jörg Stülke]]'s lab |
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
Revision as of 08:31, 3 September 2013
- Description: Enzyme I, general (non sugar-specific) component of the PTS. Enzyme I transfers the phosphoryl group from phosphoenolpyruvate (PEP) to the phosphoryl carrier protein (HPr)
| Gene name | ptsI |
| Synonyms | |
| Essential | no |
| Product | phosphotransferase system (PTS) enzyme I |
| Function | PTS-dependent sugar transport |
| Gene expression levels in SubtiExpress: ptsI | |
| Interactions involving this protein in SubtInteract: PtsI | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 62,9 kDa, 4.59 |
| Gene length, protein length | 1710 bp, 570 amino acids |
| Immediate neighbours | ptsH, splA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 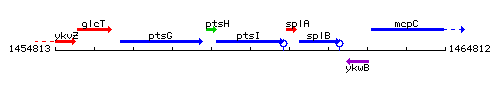
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed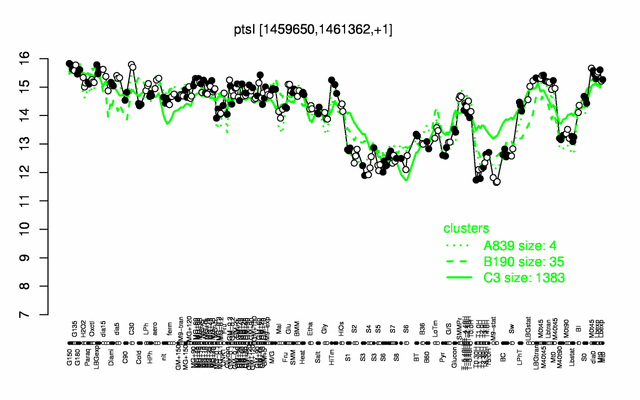
| |
Contents
Categories containing this gene/protein
phosphotransferase systems, phosphoproteins
This gene is a member of the following regulons
GlcT regulon, stringent response
The gene
Basic information
- Locus tag: BSU13910
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Phosphoenolpyruvate + protein L-histidine = pyruvate + protein N(pi)-phospho-L-histidine (according to Swiss-Prot) PEP-dependent autophosphorylation on His-189, transfer of the phosphoryl group to HPr (His-15)
- Protein family: PEP-utilizing enzyme family (according to Swiss-Prot) PEP-utilizing enzyme family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- HPr binding site (N-Terminal Domain)
- pyruvate binding site (C-Terminal Domain)
- pyrophosphate/phosphate carrier histidine (central Domain)
- Modification:
- transient autophosphorylation on His-189
- in vivo also phosphorylated on Ser-34 or Ser-36 PubMed
- Cofactor(s): Magnesium
- Effectors of protein activity:
- Localization: cytoplasm, even distribution PubMed
Database entries
- UniProt: P08838
- KEGG entry: [3]
- E.C. number: 2.7.3.9 2.7.3.9]
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- ptsG: transcriptional antitermination via the GlcT-dependent RNA switch PubMed
- Additional information:
Biological materials
- Mutant: available in Jörg Stülke's lab:
- Expression vector:
- pAG3 (His-tag) PubMed, available in Galinier lab
- for expression, purification in E. coli (His-tag), in pWH844: pGP813 available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- YFP fusion: B. subtilis GP1276 ptsI-yfp ermC, available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Josef Deutscher, Paris-Grignon, France
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References