Difference between revisions of "PtsH"
| Line 120: | Line 120: | ||
** pGP438 (with N-terminal Strep-tag, in [[pGP172]]), available in [[Stülke]] lab | ** pGP438 (with N-terminal Strep-tag, in [[pGP172]]), available in [[Stülke]] lab | ||
** pAG2 (His-tag), available in [[Anne Galinier]] lab | ** pAG2 (His-tag), available in [[Anne Galinier]] lab | ||
| − | ** pGP371( | + | ** pGP371(expression / purification of HPr-S46A, with His-tag from ''E. coli'', in [[pWH844]]), available in [[Stülke]] lab |
| + | ** pGP1415(HPr, expression in ''B. subtilis'', from [[pBQ200]]), available in [[Stülke]] lab | ||
| + | ** pGP1416(HPr-H15A, expression in ''B. subtilis'', from [[pBQ200]]), available in [[Stülke]] lab | ||
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
Revision as of 21:36, 18 March 2010
- Description: HPr, General component of the sugar phosphotransferase system (PTS).
| Gene name | ptsH |
| Synonyms | |
| Essential | no |
| Product | histidine-containing phosphocarrier protein HPr of the PTS |
| Function | PTS-dependent sugar transport and carbon catabolite repression |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism, Sugar catabolism | |
| MW, pI | 9,1 kDa, 4.58 |
| Gene length, protein length | 264 bp, 88 amino acids |
| Immediate neighbours | ptsG, ptsI |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 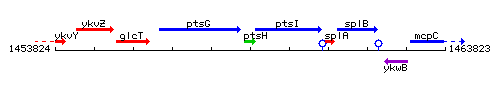
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU13900
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry:[2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein HPr N(pi)-phospho-L-histidine + protein EIIA = protein HPr + protein EIIA N(tau)-phospho-L-histidine (according to Swiss-Prot) Protein HPr N(pi)-phospho-L-histidine + protein EIIA = protein HPr + protein EIIA N(tau)-phospho-L-histidine
- Protein family: HPr domain (according to Swiss-Prot) HPr family
- Paralogous protein(s): Crh
Extended information on the protein
- Kinetic information:
- Domains: HPr Domain (2–88)
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions: HPr-LicT, HPr-SacY, HPr-SacT, HPr-GlcT, HPr-GlpK, GapA-HPr PubMed, HPr-MtlR, HPr-LicR, HPr-LevR,HPr-ManR, HPr(Ser-46)-P-CcpA PubMed, HPr-RbsR PubMed, HprK-HPr PubMed, HPr(His)-P-YesS PubMed, HPr-PtsG PubMed
- Localization: cytoplasm (according to Swiss-Prot), Cytoplasm PubMed
Database entries
- Structure: 1KKM (complex of L. casei HprK with B. subtilis HPr-Ser-P), 1KKL (complex of Lactobacillus casei HprK with B. subtilis HPr), 2HID (NMR)
- UniProt: P08877
- KEGG entry: [3]
- E.C. number: 2.7.11.-
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- ptsG: transcriptional antitermination via the GlcT-dependent RNA switch PubMed
- Additional information:
Biological materials
- Mutant: MZ303 (cat), GP507 ptsH1 (S46A), GP506 (ptsH-H15A), available in Stülke lab
- Expression vector:
- pGP438 (with N-terminal Strep-tag, in pGP172), available in Stülke lab
- pAG2 (His-tag), available in Anne Galinier lab
- pGP371(expression / purification of HPr-S46A, with His-tag from E. coli, in pWH844), available in Stülke lab
- pGP1415(HPr, expression in B. subtilis, from pBQ200), available in Stülke lab
- pGP1416(HPr-H15A, expression in B. subtilis, from pBQ200), available in Stülke lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Görke lab
- Antibody: available in Stülke lab
Labs working on this gene/protein
Josef Deutscher, Paris-Grignon, France
Jörg Stülke, University of Göttingen, Germany Homepage
Wolfgang Hillen, Erlangen University, Germany Homepage
Richard Brennan, Houston, Texas, USA Homepage
Boris Görke, University of Göttingen, Germany Homepage
Anne Galinier, University of Marseille, France
Your additional remarks
References