Difference between revisions of "PtsH"
(→Basic information/ Evolution) |
|||
| Line 20: | Line 20: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ptsG]]'', ''[[ptsI]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ptsG]]'', ''[[ptsI]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ptsH_nucleotide.txt Gene sequence (+200bp) ]''' |
| − | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ptsH_protein.txt Protein sequence]''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:ptsH_context.gif]] |
|- | |- | ||
|} | |} | ||
Revision as of 14:54, 12 January 2009
- Description: General component of the sugar phosphotransferase system (PTS).
| Gene name | ptsH |
| Synonyms | |
| Essential | no |
| Product | histidine-containing phosphocarrier protein of the PTS |
| Function | mediates carbon catabolite repression (CCR) |
| MW, pI | 9,1 kDa, 4.58 |
| Gene length, protein length | 264 bp, 88 amino acids |
| Immediate neighbours | ptsG, ptsI |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 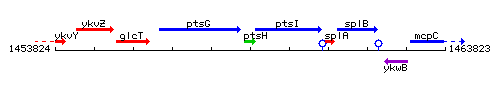
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry:[2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein HPr N(pi)-phospho-L-histidine + protein EIIA = protein HPr + protein EIIA N(tau)-phospho-L-histidine
- Protein family: HPr family
- Paralogous protein(s): Crh
Extended information on the protein
- Kinetic information:
- Domains: HPr Domain (2–88)
- Modification: phosphorylation (Ser-12)
- Cofactor(s):
- Effectors of protein activity:
- Interactions: GapA-HPr PubMed, HPr-MtlR, HPr-LicR, HPr-LevR,HPr-ManR, YesS-HPr (HPr-His-P), HPr-CcpA PubMed, HPr-RbsR PubMed, HprK-HPr PubMed
- Localization: Cytoplasm
Database entries
- Structure: NCBI, complex of L. Casei HprK with B. Subtilis HPr NCBI, complex of L. Casei HprK with B. Subtilis HPr-Ser-P NCBI
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: [5]
Additional information
Expression and regulation
- Additional information:
Biological materials
Labs working on this gene/protein
Josef Deutscher, Paris-Grignon, France
Jörg Stülke, University of Göttingen, Germany Homepage
Wolfgang Hillen, Erlangen University, Germany Homepage
Richard Brennan, Houston, Texas, USA Homepage
Boris Görke, University of Göttingen, Germany Homepage
Anne Galinier, University of Marseille, France
Your additional remarks
References
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed
- Macek B, Mijakovic I, Olsen JV (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics 6(4): 697-707. PubMed
- Mahr K, Esteban CD, Hillen W (2002) Cross communication between components of carbon catabolite repression of Lactobacillus casei and Bacillus megaterium. J Mol Microbiol Biotechnol. 4(5): 489-94. PubMed
- Juy M, Penin F, Favier A, Galinier A (2003) Dimerization of Crh by reversible 3D domain swapping induces structural adjustments to its monomeric homologue Hpr J Mol Biol. 332(4): 767-76. PubMed
- Müller W, Horstmann N, Hillen W (2006) The transcription regulator RbsR represents a novel interaction partner of the phosphoprotein HPr-Ser46-P in Bacillus subtilis FEBS J. 273(6): 1251-61. PubMed
- Lavergne JP, Jault JM, Galinier A (2002) Insights into the functioning of Bacillus subtilis HPr kinase/phosphatase: affinity for its protein substrates and role of cations and phosphate Biochemistry 41(20): 6218-25. PubMed