Difference between revisions of "PtpZ"
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ugd]]'', ''[[ptkA]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ugd]]'', ''[[ptkA]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU36240 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU36240 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU36240 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU36240 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU36240 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU36240 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ywqE_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ywqE_context.gif]] | ||
Revision as of 11:27, 14 May 2013
- Description: phosphotyrosine protein phosphatase, antagonist to PtkA
| Gene name | ptpZ |
| Synonyms | ywqE |
| Essential | no |
| Product | phosphotyrosine protein phosphatase, antagonist to PtkA |
| Function | protein tyrosine dephosphorylation |
| Gene expression levels in SubtiExpress: ptpZ | |
| Interactions involving this protein in SubtInteract: PtpZ | |
| MW, pI | 28 kDa, 6.66 |
| Gene length, protein length | 762 bp, 254 aa |
| Immediate neighbours | ugd, ptkA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 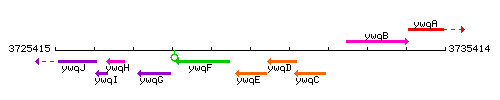
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed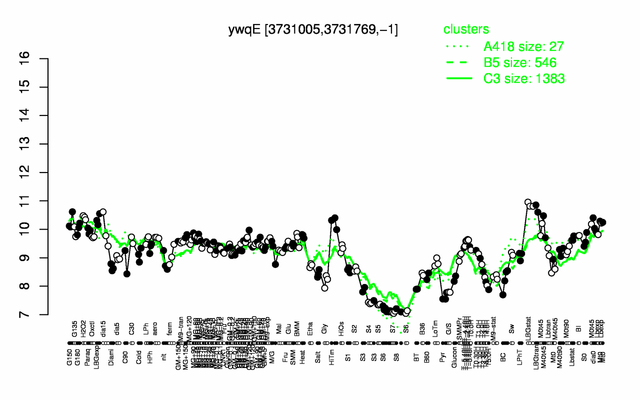
| |
Contents
Categories containing this gene/protein
biofilm formation, protein modification
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU36240
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein tyrosine phosphate + H2O = protein tyrosine + phosphate (according to Swiss-Prot), dephosphorylation of Ugd, TuaD and PtkA PubMed
- Protein family: cpsB/capC family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P96717
- KEGG entry: [2]
- E.C. number: 3.1.3.48
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Hyoun Sook Kim, Sang Jae Lee, Hye Jin Yoon, Doo Ri An, Do Jin Kim, Soon-Jong Kim, Se Won Suh
Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J Struct Biol: 2011, 175(3);442-50
[PubMed:21605684]
[WorldCat.org]
[DOI]
(I p)
Taryn B Kiley, Nicola R Stanley-Wall
Post-translational control of Bacillus subtilis biofilm formation mediated by tyrosine phosphorylation.
Mol Microbiol: 2010, 78(4);947-63
[PubMed:20815827]
[WorldCat.org]
[DOI]
(I p)
Ivan Mijakovic, Lucia Musumeci, Lutz Tautz, Dina Petranovic, Robert A Edwards, Peter Ruhdal Jensen, Tomas Mustelin, Josef Deutscher, Nunzio Bottini
In vitro characterization of the Bacillus subtilis protein tyrosine phosphatase YwqE.
J Bacteriol: 2005, 187(10);3384-90
[PubMed:15866923]
[WorldCat.org]
[DOI]
(P p)
Ivan Mijakovic, Sandrine Poncet, Grégory Boël, Alain Mazé, Sylvie Gillet, Emmanuel Jamet, Paulette Decottignies, Christophe Grangeasse, Patricia Doublet, Pierre Le Maréchal, Josef Deutscher
Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases.
EMBO J: 2003, 22(18);4709-18
[PubMed:12970183]
[WorldCat.org]
[DOI]
(P p)