Difference between revisions of "PtkA"
| Line 54: | Line 54: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' BY-kinase |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 64: | Line 64: | ||
* '''Domains:''' | * '''Domains:''' | ||
| − | * '''Modification:''' | + | * '''Modification:''' autophosphorylation at residues Y225, Y227 and Y228 (primary site) |
| − | * '''Cofactor(s):''' | + | * '''Cofactor(s):''' ATP |
| − | * '''Effectors of protein activity:''' | + | * '''Effectors of protein activity:''' TkmA - transmembrane modulator, activates PtkA autophosphorylation and substrate phosphorylation |
* '''Interactions:''' | * '''Interactions:''' | ||
| Line 100: | Line 100: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' | + | * '''Mutant:''' KO strain created with pMUTIN-2 |
| − | * '''Expression vector:''' | + | * '''Expression vector:''' pQE-30, N-terminally 6xHis-tagged |
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| Line 112: | Line 112: | ||
* '''Antibody:''' | * '''Antibody:''' | ||
| − | =Labs working on this gene/protein= | + | =Labs working on this gene/protein= Ivan Mijakovic |
=Your additional remarks= | =Your additional remarks= | ||
| Line 119: | Line 119: | ||
# Mijakovic I, Poncet S, Boël G, Mazé A, Gillet S, Decottignies P, Grangeasse C, Jamet E, Doublet P, Le Maréchal P, Deutscher J (2003) Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J 22:4709-4718. [http://www.ncbi.nlm.nih.gov/sites/entrez/12970183 PubMed] | # Mijakovic I, Poncet S, Boël G, Mazé A, Gillet S, Decottignies P, Grangeasse C, Jamet E, Doublet P, Le Maréchal P, Deutscher J (2003) Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J 22:4709-4718. [http://www.ncbi.nlm.nih.gov/sites/entrez/12970183 PubMed] | ||
| − | # Mijakovic I, Petranovic D, Macek B, Cepo T, Mann M, Davies J, Jensen PR, Vujaklija D | + | # Mijakovic I, Petranovic D, Deutscher J (2004) How tyrosine phosphorylation affects the UDP-glucose dehydrogenase activity of Bacillus subtilis YwqF. J Mol Microbiol Biotechnol 8:19-25. [http://www.ncbi.nlm.nih.gov/sites/entrez/15741737 PubMed] |
| + | # Mijakovic I, Petranovic D, Macek B, Cepo T, Mann M, Davies J, Jensen PR, Vujaklija D (2006) Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucl Acids Res 34:1588-1596. [http://www.ncbi.nlm.nih.gov/sites/entrez/16549871 PubMed] | ||
| + | # Petranovic D, Michelsen O, Zahradka K, Silva C, Petranovic M, Jensen PR, Mijakovic I (2007) Bacillus subtilis strain deficient for the protein-tyrosine kinase PtkA exhibits impaired DNA replication. Mol Microbiol 63:1797-1805. [http://www.ncbi.nlm.nih.gov/sites/entrez/17367396 PubMed] | ||
| + | |||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 11:20, 15 March 2009
- Description: write here
| Gene name | ptkA |
| Synonyms | ywqD |
| Essential | no |
| Product | |
| Function | unknown |
| MW, pI | 25 kDa, 9.628 |
| Gene length, protein length | 711 bp, 237 aa |
| Immediate neighbours | ptpZ, tkmA |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 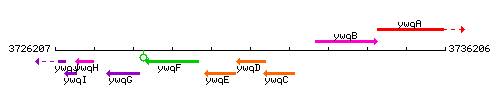
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: BY-kinase
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: autophosphorylation at residues Y225, Y227 and Y228 (primary site)
- Cofactor(s): ATP
- Effectors of protein activity: TkmA - transmembrane modulator, activates PtkA autophosphorylation and substrate phosphorylation
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry:
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: KO strain created with pMUTIN-2
- Expression vector: pQE-30, N-terminally 6xHis-tagged
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
=Labs working on this gene/protein= Ivan Mijakovic
Your additional remarks
References
- Mijakovic I, Poncet S, Boël G, Mazé A, Gillet S, Decottignies P, Grangeasse C, Jamet E, Doublet P, Le Maréchal P, Deutscher J (2003) Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J 22:4709-4718. PubMed
- Mijakovic I, Petranovic D, Deutscher J (2004) How tyrosine phosphorylation affects the UDP-glucose dehydrogenase activity of Bacillus subtilis YwqF. J Mol Microbiol Biotechnol 8:19-25. PubMed
- Mijakovic I, Petranovic D, Macek B, Cepo T, Mann M, Davies J, Jensen PR, Vujaklija D (2006) Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucl Acids Res 34:1588-1596. PubMed
- Petranovic D, Michelsen O, Zahradka K, Silva C, Petranovic M, Jensen PR, Mijakovic I (2007) Bacillus subtilis strain deficient for the protein-tyrosine kinase PtkA exhibits impaired DNA replication. Mol Microbiol 63:1797-1805. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed