Difference between revisions of "Psd"
| Line 32: | Line 32: | ||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| + | |||
| + | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[biosynthesis of lipids]]}}, | ||
| + | {{SubtiWiki category|[[cell envelope stress proteins (controlled by SigM, W, X, Y)]]}} | ||
| + | |||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[SigX regulon]]}} | ||
=The gene= | =The gene= | ||
| Line 51: | Line 58: | ||
| − | + | ||
| − | |||
| − | |||
=The protein= | =The protein= | ||
Revision as of 16:20, 8 December 2010
- Description: phosphatidylserine decarboxylase
| Gene name | psd |
| Synonyms | |
| Essential | no |
| Product | phosphatidylserine decarboxylase |
| Function | biosynthesis of phospholipids |
| Metabolic function and regulation of this protein in SubtiPathways: Lipid synthesis | |
| MW, pI | 29 kDa, 7.27 |
| Gene length, protein length | 789 bp, 263 aa |
| Immediate neighbours | ybfM, ybfN |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 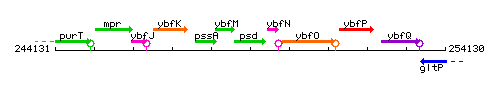
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
biosynthesis of lipids, cell envelope stress proteins (controlled by SigM, W, X, Y)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU02290
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Phosphatidyl-L-serine = phosphatidylethanolamine + CO2 (according to Swiss-Prot)
- Protein family: Type 1 subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P39822
- KEGG entry: [3]
- E.C. number: 4.1.1.65
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Letal I Salzberg, John D Helmann
Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition.
J Bacteriol: 2008, 190(23);7797-807
[PubMed:18820022]
[WorldCat.org]
[DOI]
(I p)
Min Cao, John D Helmann
The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides.
J Bacteriol: 2004, 186(4);1136-46
[PubMed:14762009]
[WorldCat.org]
[DOI]
(P p)
K Matsumoto, M Okada, Y Horikoshi, H Matsuzaki, T Kishi, M Itaya, I Shibuya
Cloning, sequencing, and disruption of the Bacillus subtilis psd gene coding for phosphatidylserine decarboxylase.
J Bacteriol: 1998, 180(1);100-6
[PubMed:9422599]
[WorldCat.org]
[DOI]
(P p)