Difference between revisions of "PrsA"
| Line 1: | Line 1: | ||
| − | * '''Description:''' post-translocational folding of exported proteins (post-translocation molecular chaperone) <br/><br/> | + | * '''Description:''' lipoprotein, post-translocational folding of exported proteins (post-translocation molecular chaperone) <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
| Line 38: | Line 38: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 74: | Line 70: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' catalyses the post-translocational folding of exported proteins {{PubMed|20487272}}, required for folding of [[penicillin-binding proteins]] ([[PbpA]], [[PbpB]], [[PbpC]], [[PbpD]]) {{PubMed|20487272}} | + | * '''Catalyzed reaction/ biological activity:''' |
| + | ** catalyses the post-translocational folding of exported proteins {{PubMed|20487272}}, required for folding of [[penicillin-binding proteins]] ([[PbpA]], [[PbpB]], [[PbpC]], [[PbpD]]) {{PubMed|20487272}} | ||
* '''Protein family:''' PpiC domain (according to Swiss-Prot) | * '''Protein family:''' PpiC domain (according to Swiss-Prot) | ||
| Line 84: | Line 81: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
** PrsA is a lipoprotein with a N-acetyl-S-diacyl-glyceryl-cysteine structure {{PubMed|22303020}} | ** PrsA is a lipoprotein with a N-acetyl-S-diacyl-glyceryl-cysteine structure {{PubMed|22303020}} | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 118: | Line 115: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=prsA_1070364_1071242_-1 prsA] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=prsA_1070364_1071242_-1 prsA] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 125: | Line 122: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** [[PrsA]] is subject to degradation by [[WprA]] and other extracellular proteases {{PubMed|24362423}} | ||
=Biological materials = | =Biological materials = | ||
| Line 146: | Line 144: | ||
=References= | =References= | ||
| − | <pubmed>12634326,14976191,11807061,10096076,18763711, 10871614 20487272 22303020</pubmed> | + | <pubmed>12634326,14976191,11807061,10096076,18763711, 10871614 20487272 22303020 24362423 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:28, 16 January 2014
- Description: lipoprotein, post-translocational folding of exported proteins (post-translocation molecular chaperone)
| Gene name | prsA |
| Synonyms | |
| Essential | yes PubMed |
| Product | post-translocation molecular chaperone |
| Function | protein folding |
| Gene expression levels in SubtiExpress: prsA | |
| Interactions involving this protein in SubtInteract: PrsA | |
| Metabolic function and regulation of this protein in SubtiPathways: Protein secretion | |
| MW, pI | 32 kDa, 9.122 |
| Gene length, protein length | 876 bp, 292 aa |
| Immediate neighbours | yhaL, sscA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 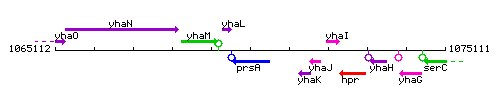
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
protein secretion, essential genes, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU09950
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: PpiC domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- PrsA is a lipoprotein with a N-acetyl-S-diacyl-glyceryl-cysteine structure PubMed
- Effectors of protein activity:
- Interactions:
- dimeric or oligomeric protein PubMed
Database entries
- Structure: 1ZK6
- UniProt: P24327
- KEGG entry: [2]
- E.C. number: 5.2.1.8
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Laxmi Krishnappa, Carmine G Monteferrante, Jolanda Neef, Annette Dreisbach, Jan Maarten van Dijl
Degradation of extracytoplasmic catalysts for protein folding in Bacillus subtilis.
Appl Environ Microbiol: 2014, 80(4);1463-8
[PubMed:24362423]
[WorldCat.org]
[DOI]
(I p)
Kenji Kurokawa, Kyoung-Hwa Ryu, Rie Ichikawa, Akiko Masuda, Min-Su Kim, Hanna Lee, Jun-Ho Chae, Takashi Shimizu, Tatsuya Saitoh, Koichi Kuwano, Shizuo Akira, Naoshi Dohmae, Hiroshi Nakayama, Bok Luel Lee
Novel bacterial lipoprotein structures conserved in low-GC content gram-positive bacteria are recognized by Toll-like receptor 2.
J Biol Chem: 2012, 287(16);13170-81
[PubMed:22303020]
[WorldCat.org]
[DOI]
(I p)
Hanne-Leena Hyyryläinen, Bogumila C Marciniak, Kathleen Dahncke, Milla Pietiäinen, Pascal Courtin, Marika Vitikainen, Raili Seppala, Andreas Otto, Dörte Becher, Marie-Pierre Chapot-Chartier, Oscar P Kuipers, Vesa P Kontinen
Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis.
Mol Microbiol: 2010, 77(1);108-27
[PubMed:20487272]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Marika Vitikainen, Ilkka Lappalainen, Raili Seppala, Haike Antelmann, Harry Boer, Suvi Taira, Harri Savilahti, Michael Hecker, Mauno Vihinen, Matti Sarvas, Vesa P Kontinen
Structure-function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C-terminal domains in protein folding and secretion in Bacillus subtilis.
J Biol Chem: 2004, 279(18);19302-14
[PubMed:14976191]
[WorldCat.org]
[DOI]
(P p)
Eva Wahlström, Marika Vitikainen, Vesa P Kontinen, Matti Sarvas
The extracytoplasmic folding factor PrsA is required for protein secretion only in the presence of the cell wall in Bacillus subtilis.
Microbiology (Reading): 2003, 149(Pt 3);569-577
[PubMed:12634326]
[WorldCat.org]
[DOI]
(P p)
Tiina Pummi, Soile Leskelä, Eva Wahlström, Ulf Gerth, Harold Tjalsma, Michael Hecker, Matti Sarvas, Vesa P Kontinen
ClpXP protease regulates the signal peptide cleavage of secretory preproteins in Bacillus subtilis with a mechanism distinct from that of the Ecs ABC transporter.
J Bacteriol: 2002, 184(4);1010-8
[PubMed:11807061]
[WorldCat.org]
[DOI]
(P p)
H L Hyyrylainen, M Vitikainen, J Thwaite, H Wu, M Sarvas, C R Harwood, V P Kontinen, K Stephenson
D-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis.
J Biol Chem: 2000, 275(35);26696-703
[PubMed:10871614]
[WorldCat.org]
[DOI]
(P p)
S Leskelä, E Wahlström, V P Kontinen, M Sarvas
Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the Lgt gene.
Mol Microbiol: 1999, 31(4);1075-85
[PubMed:10096076]
[WorldCat.org]
[DOI]
(P p)