Difference between revisions of "PrpC"
| Line 16: | Line 16: | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU15760 prpC] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU15760 prpC] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/ | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=prpC prpC]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 27 kDa, 4.355 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 27 kDa, 4.355 | ||
Revision as of 10:45, 7 January 2014
- Description: protein phosphatase
| Gene name | prpC |
| Synonyms | yloO |
| Essential | no |
| Product | protein phosphatase |
| Function | antagonist of PrkC-dependent phosphorylation |
| Gene expression levels in SubtiExpress: prpC | |
| Metabolic function and regulation of this protein in SubtiPathways: prpC | |
| MW, pI | 27 kDa, 4.355 |
| Gene length, protein length | 762 bp, 254 aa |
| Immediate neighbours | yloN, prkC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 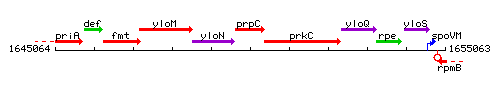
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed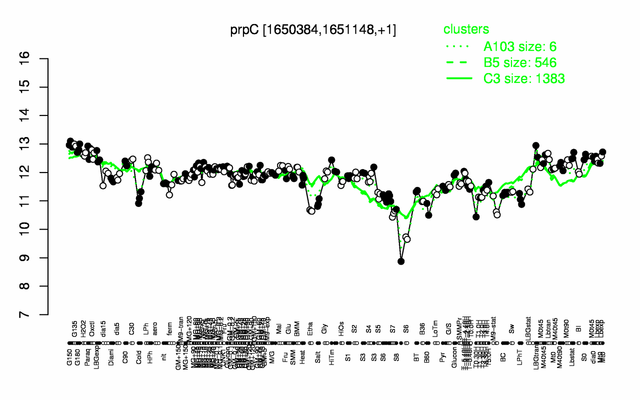
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15760
Phenotypes of a mutant
A prpC mutant is less lytic in late stationary phase. PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: A phosphoprotein + H2O = a protein + phosphate (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Proteins dephosphorylated by PrpC
CpgA, EF-Tu, YezB PubMed, HPr PubMed
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): divalent cations such as magnesium or manganese
- Effectors of protein activity: inhibited by inorganic phosphate and glycero-2-phosphate PubMed
Database entries
- UniProt: O34779
- KEGG entry: [2]
- E.C. number: 3.1.3.16
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: OMG401 (aphA3), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Gunjan Arora, Andaleeb Sajid, Mary Diana Arulanandh, Richa Misra, Anshika Singhal, Santosh Kumar, Lalit K Singh, Abid R Mattoo, Rishi Raj, Souvik Maiti, Sharmila Basu-Modak, Yogendra Singh
Zinc regulates the activity of kinase-phosphatase pair (BasPrkC/BasPrpC) in Bacillus anthracis.
Biometals: 2013, 26(5);715-30
[PubMed:23793375]
[WorldCat.org]
[DOI]
(I p)
Cédric Absalon, Michal Obuchowski, Edwige Madec, Delphine Delattre, I Barry Holland, Simone J Séror
CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 3);932-943
[PubMed:19246764]
[WorldCat.org]
[DOI]
(P p)
Kalpana D Singh, Matthias H Schmalisch, Jörg Stülke, Boris Görke
Carbon catabolite repression in Bacillus subtilis: quantitative analysis of repression exerted by different carbon sources.
J Bacteriol: 2008, 190(21);7275-84
[PubMed:18757537]
[WorldCat.org]
[DOI]
(I p)
Kalpana D Singh, Sven Halbedel, Boris Görke, Jörg Stülke
Control of the phosphorylation state of the HPr protein of the phosphotransferase system in Bacillus subtilis: implication of the protein phosphatase PrpC.
J Mol Microbiol Biotechnol: 2007, 13(1-3);165-71
[PubMed:17693724]
[WorldCat.org]
[DOI]
(P p)
Adam Iwanicki, Krzysztof Hinc, Simone Seror, Grzegorz Wegrzyn, Michal Obuchowski
Transcription in the prpC-yloQ region in Bacillus subtilis.
Arch Microbiol: 2005, 183(6);421-30
[PubMed:16025310]
[WorldCat.org]
[DOI]
(P p)
Kristi E Pullen, Ho-Leung Ng, Pei-Yi Sung, Matthew C Good, Stephen M Smith, Tom Alber
An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-Family Ser/Thr protein phosphatase.
Structure: 2004, 12(11);1947-54
[PubMed:15530359]
[WorldCat.org]
[DOI]
(P p)
Tatiana A Gaidenko, Tae-Jong Kim, Chester W Price
The PrpC serine-threonine phosphatase and PrkC kinase have opposing physiological roles in stationary-phase Bacillus subtilis cells.
J Bacteriol: 2002, 184(22);6109-14
[PubMed:12399479]
[WorldCat.org]
[DOI]
(P p)
M Obuchowski, E Madec, D Delattre, G Boël, A Iwanicki, D Foulger, S J Séror
Characterization of PrpC from Bacillus subtilis, a member of the PPM phosphatase family.
J Bacteriol: 2000, 182(19);5634-8
[PubMed:10986276]
[WorldCat.org]
[DOI]
(P p)