Difference between revisions of "PnbA"
(→Biological materials) |
|||
| Line 85: | Line 85: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1C7J 1C7J] | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1C7J 1C7J] {{PubMed|10535917}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P37967 P37967] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P37967 P37967] | ||
| Line 131: | Line 131: | ||
=References= | =References= | ||
| − | <pubmed>7828905,21574267,17218307, </pubmed> | + | <pubmed>7828905,21574267,17218307, 10535917 22127613 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:18, 2 December 2011
- Description: para-nitrobenzyl esterase
| Gene name | pnbA |
| Synonyms | estB |
| Essential | no |
| Product | para-nitrobenzyl esterase |
| Function | lipid degradation |
| MW, pI | 53 kDa, 4.776 |
| Gene length, protein length | 1467 bp, 489 aa |
| Immediate neighbours | slrR, padC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 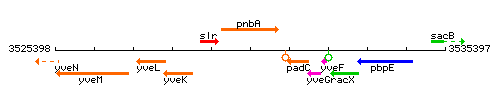
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
utilization of lipids, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU34390
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Triacylglycerol + H2O = diacylglycerol + a carboxylate (according to Swiss-Prot)
- Protein family: type-B carboxylesterase/lipase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Ser-189 PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P37967
- KEGG entry: [2]
- E.C. number: 3.1.1.3 3.1.1.1]
Additional information
Expression and regulation
- Operon:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Xiaozhen Yu, Sara C Sigler, Delwar Hossain, Monika Wierdl, Steven R Gwaltney, Philip M Potter, Randy M Wadkins
Global and local molecular dynamics of a bacterial carboxylesterase provide insight into its catalytic mechanism.
J Mol Model: 2012, 18(6);2869-83
[PubMed:22127613]
[WorldCat.org]
[DOI]
(I p)
Doris Ribitsch, Sonja Heumann, Eva Trotscha, Enrique Herrero Acero, Katrin Greimel, Regina Leber, Ruth Birner-Gruenberger, Sigrid Deller, Inge Eiteljoerg, Peter Remler, Thomas Weber, Petra Siegert, Karl-Heinz Maurer, Ilaria Donelli, Giuliano Freddi, Helmut Schwab, Georg M Guebitz
Hydrolysis of polyethyleneterephthalate by p-nitrobenzylesterase from Bacillus subtilis.
Biotechnol Prog: 2011, 27(4);951-60
[PubMed:21574267]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
B Spiller, A Gershenson, F H Arnold, R C Stevens
A structural view of evolutionary divergence.
Proc Natl Acad Sci U S A: 1999, 96(22);12305-10
[PubMed:10535917]
[WorldCat.org]
[DOI]
(P p)
J Zock, C Cantwell, J Swartling, R Hodges, T Pohl, K Sutton, P Rosteck, D McGilvray, S Queener
The Bacillus subtilis pnbA gene encoding p-nitrobenzyl esterase: cloning, sequence and high-level expression in Escherichia coli.
Gene: 1994, 151(1-2);37-43
[PubMed:7828905]
[WorldCat.org]
[DOI]
(P p)