Difference between revisions of "Pgi"
| Line 28: | Line 28: | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:pgi_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:pgi_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=pgi_3220731_3222083_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:pgi_expression.png|500px]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | |||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
Revision as of 08:07, 23 April 2012
- Description: glucose 6-phosphate isomerase, glycolytic / gluconeogenic enzyme
| Gene name | pgi |
| Synonyms | yugL |
| Essential | no |
| Product | glucose-6-phosphate isomerase |
| Function | enzyme in glycolysis / gluconeogenesis |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism, Sugar catabolism | |
| MW, pI | 50.4 kDa, 4.85 |
| Gene length, protein length | 1353 bp, 451 amino acids |
| Immediate neighbours | yugM, yugK |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 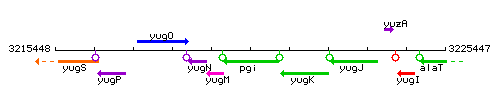
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed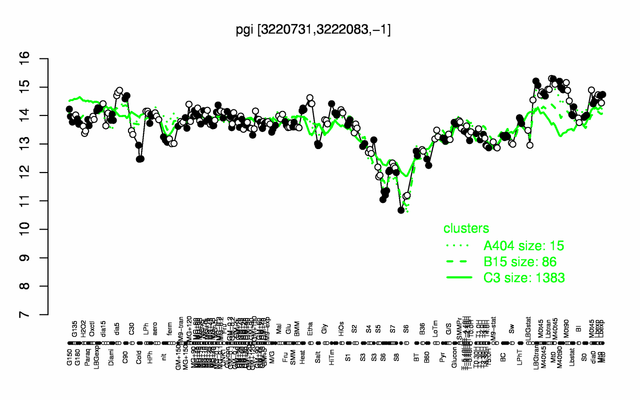
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU31350
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: D-glucose 6-phosphate = D-fructose 6-phosphate (according to Swiss-Prot) D-glucose 6-phosphate = D-fructose 6-phosphate
- Protein family: GPI family (according to Swiss-Prot) GPI family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Reversible Michaelis-Menten PubMed
- Domains:
- Modification: phosphorylation on Thr-39 PubMed
- Cofactor(s):
- Effectors of protein activity: competitively inhibited by 6-phosphogluconate (in B.caldotenax, B.stearothermophilus) PubMed
- Localization:
- cytoplasm (according to Swiss-Prot), cytoplasm
Database entries
- UniProt: P80860
- KEGG entry: [3]
- E.C. number: 5.3.1.9
Additional information
- extensive information on the structure and enzymatic properties of Pgi can be found at Proteopedia
Expression and regulation
- Sigma factor:
- Regulation: constitutively expressed PubMed
- Additional information:
Biological materials
- Mutant: GP508 (spc), available in Stülke lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany homepage
Your additional remarks
References
Yian Liang Lee, TienHsiung Thomas Li
Crystallization and preliminary crystallographic study of the phosphoglucose isomerase from Bacillus subtilis.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2008, 64(Pt 12);1181-3
[PubMed:19052382]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
J Stülke, I Martin-Verstraete, P Glaser, G Rapoport
Characterization of glucose-repression-resistant mutants of Bacillus subtilis: identification of the glcR gene.
Arch Microbiol: 2001, 175(6);441-9
[PubMed:11491085]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
J Y Lin, C Prasad
Selection of a mutant of Bacillus subtilis deficient in glucose-6-phosphate dehydrogenase and phosphoglucoisomerase.
J Gen Microbiol: 1974, 83(2);419-21
[PubMed:4214896]
[WorldCat.org]
[DOI]
(P p)
C Prasad, E Freese
Cell lysis of Bacillus subtilis caused by intracellular accumulation of glucose-1-phosphate.
J Bacteriol: 1974, 118(3);1111-22
[PubMed:4275311]
[WorldCat.org]
[DOI]
(P p)