Difference between revisions of "PfkA"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || catabolic enzyme in glycolysis | |style="background:#ABCDEF;" align="center"|'''Function''' || catabolic enzyme in glycolysis | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/PfkA PfkA] | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism], [http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism], [http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism]''' | ||
Revision as of 13:22, 28 July 2011
- Description: phosphofructokinase, glycolytic enzyme
| Gene name | pfkA |
| Synonyms | pfk |
| Essential | no |
| Product | 6-phosphofructokinase |
| Function | catabolic enzyme in glycolysis |
| Interactions involving this protein in SubtInteract: PfkA | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism, Sugar catabolism | |
| MW, pI | 34,1 kDa, 6.14 |
| Gene length, protein length | 957 bp, 319 amino acids |
| Immediate neighbours | pyk, accA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 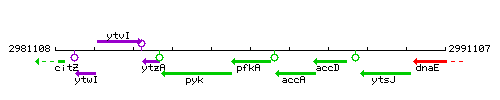
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29190
Phenotypes of a mutant
- essential according to Kobayashi et al., dispensable according to Munoz-Marquez and Ponce-Rivas
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + D-fructose 6-phosphate = ADP + D-fructose 1,6-bisphosphate (according to Swiss-Prot)
- Protein family: phosphofructokinase family (according to Swiss-Prot) phosphofructokinase family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Allosteric Regulation (Reversible) PubMed
- Domains:
- 3 x nucleotide binding domain (ATP) (21–25), (154–158), (171–187)
- Modification:
- Cofactor(s): Mg2+
- Effectors of protein activity:
- Inhibited by citrate, PEP (Hill Coefficient 3) and Ca2+ (competes with Mg2+) in B. licheniformes PubMed.
- Inhibited by ATP (competitively) and f6p (non-competitively) in G. stearothermophillus PubMed
- Activated by GDP and ADP in lower concentrations (1mM); above that inhibition, competing with the ATP for the binding site (in G. stearothermophillus) PubMed
- Activated by NH4+ PubMed
- Interactions: part of the RNA degradosome
- Localization: cytoplasm (Homogeneous) PubMed
Database entries
- Structure: 1MTO (mutant, complex with fructose-6-phosphate, Geobacillus stearothermophilus), 4PFK (Geobacillus stearothermophilus), Geobacillus stearothermophilus NCBI, Mutant form, complex with fructose-6-phosphate Geobacillus stearothermophilus NCBI
- UniProt: O34529
- KEGG entry: [3]
- E.C. number: 2.7.1.11
Additional information
- PfkA is a moonlighting protein. PubMed
- extensive information on the structure and enzymatic properties of PfkA can be found at Proteopedia
Expression and regulation
- Sigma factor:
- Regulation:
- twofold induced by glucose PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- for expression/ purification from B. subtilis with N-terminal Strep-tag, for SPINE, in pGP380: pGP87, available in Stülke lab
- for expression/ purification from B. subtilis with C-terminal Strep-tag, for SPINE, in pGP382: pGP1266, available in Stülke lab
- for expression/ purification from E. coli with N-terminal His-tag, in pWH844: pGP393, available in Stülke lab
- for expression in B. subtilis, in pBQ200: pGP1422, available in Stülke lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References