Difference between revisions of "PfkA"
(→Extended information on the protein) |
|||
| Line 75: | Line 75: | ||
* '''Interactions:''' [[PfkA]]-[[Pgm]], [[PfkA]]-[[Eno]], [[PfkA]]-[[Rny]], [[PfkA]]-[[PnpA]], [[PfkA]]-[[RnjA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/19193632 PubMed] | * '''Interactions:''' [[PfkA]]-[[Pgm]], [[PfkA]]-[[Eno]], [[PfkA]]-[[Rny]], [[PfkA]]-[[PnpA]], [[PfkA]]-[[RnjA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/19193632 PubMed] | ||
| − | * '''Localization:''' Cytoplasm (Homogeneous) | + | * '''Localization:''' Cytoplasm (Homogeneous) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] |
=== Database entries === | === Database entries === | ||
Revision as of 14:19, 24 March 2009
- Description: phosphofructokinase, glycolytic enzyme
| Gene name | pfkA |
| Synonyms | pfk |
| Essential | yes |
| Product | 6-phosphofructokinase |
| Function | catabolic enzyme in glycolysis |
| MW, pI | 34,1 kDa, 6.14 |
| Gene length, protein length | 957 bp, 319 amino acids |
| Immediate neighbours | accA, pyk |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 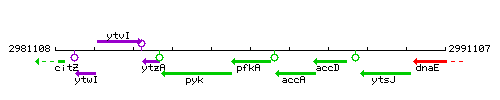
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates: 2985630 - 2986586
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + D-fructose 6-phosphate = ADP + D-fructose 1,6-bisphosphate
- Protein family: phosphofructokinase family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- 3 x nucleotide binding domain (ATP) (21–25), (154–158), (171–187)
- Modification:
- Cofactor(s): ATP
- Effectors of protein activity: inhibited by citrate, ATP, PEP, Ca2+, and others (in B. licheniformes) PubMed
- Localization: Cytoplasm (Homogeneous) PubMed
Database entries
- Structure: Geobacillus stearothermophilus NCBI, Mutant form, complex with fructose-6-phosphate Geobacillus stearothermophilus NCBI
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: [5]
Additional information
Expression and regulation
- Sigma factor:
- Regulation: twofold induced by glucose PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector: pGP393 (N-terminal His-tag, in pWH844), pGP87 (N-terminal Strep-tag, for SPINE, expression in B. subtilis, in pGP380), available in Stülke lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
- Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory Proteomics 6: 2135-2146. PubMed
- Commichau, F. M., Rothe, F. M., Herzberg, C., Wagner, E., Hellwig, D., Lehnik-Habrink, M., Hammer, E., Völker, U. & Stülke, J. (2009) Novel activities of glycolytic enzymes in Bacillus subtilis: Interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteomics in press PubMed
- Ludwig, H., Homuth, G., Schmalisch, M., Dyka, F. M., Hecker, M., and Stülke, J. (2001) Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol Microbiol 41, 409-422. PubMed