Difference between revisions of "PelC"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || degradation of polygalacturonic acid | |style="background:#ABCDEF;" align="center"|'''Function''' || degradation of polygalacturonic acid | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU34950 pelC] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 24 kDa, 9.14 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 24 kDa, 9.14 | ||
Revision as of 16:23, 7 August 2012
- Description: pectate lyase C
| Gene name | pelC |
| Synonyms | yvpA |
| Essential | no |
| Product | pectate lyase C |
| Function | degradation of polygalacturonic acid |
| Gene expression levels in SubtiExpress: pelC | |
| MW, pI | 24 kDa, 9.14 |
| Gene length, protein length | 663 bp, 221 aa |
| Immediate neighbours | yvpB, yvoF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed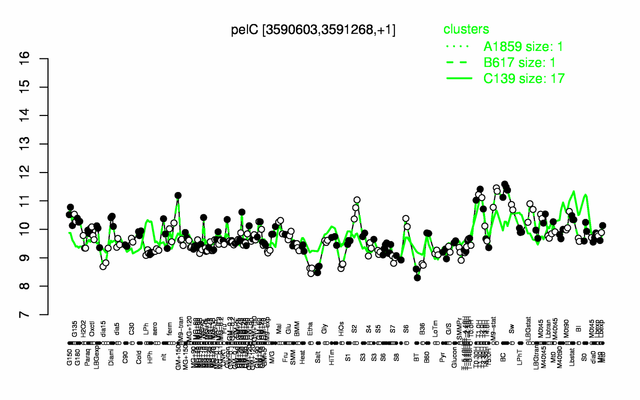
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU34950
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Eliminative cleavage of (1->4)-alpha-D-galacturonan to give oligosaccharides with 4-deoxy-alpha-D-galact-4-enuronosyl groups at their non-reducing ends (according to Swiss-Prot)
- Protein family: polysaccharide lyase 3 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- extracellular (signal peptide) PubMed
Database entries
- Structure:
- UniProt: O34310
- KEGG entry: [2]
- E.C. number: 4.2.2.2
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Margarita Soriano, Pilar Diaz, Francisco I Javier Pastor
Pectate lyase C from Bacillus subtilis: a novel endo-cleaving enzyme with activity on highly methylated pectin.
Microbiology (Reading): 2006, 152(Pt 3);617-625
[PubMed:16514142]
[WorldCat.org]
[DOI]
(P p)