Difference between revisions of "PdxT"
| Line 63: | Line 63: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 90: | Line 87: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| − | ** [[PdxS]]-[[PdxT]] | + | ** [[PdxS]]-[[PdxT]] {{PubMed|25473090,14762015,16030023,17159152}} |
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| Line 149: | Line 146: | ||
=References= | =References= | ||
| − | <pubmed>15911615,14762015,14651647 16030023, 19152323,17159152 23832367 </pubmed> | + | <pubmed>15911615,14762015,14651647 16030023, 19152323,17159152 23832367 25473090</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:27, 5 December 2014
- Description: pyridoxal-5'-phosphate synthase (glutaminase domain)
| Gene name | pdxT |
| Synonyms | yaaE |
| Essential | no |
| Product | pyridoxal-5'-phosphate synthase (glutaminase domain) |
| Function | pyridoxal-5'-phosphate biosynthesis |
| Gene expression levels in SubtiExpress: pdxT | |
| Interactions involving this protein in SubtInteract: PdxT | |
| Metabolic function and regulation of this protein in SubtiPathways: PdxT | |
| MW, pI | 21 kDa, 4.984 |
| Gene length, protein length | 588 bp, 196 aa |
| Immediate neighbours | pdxS, serS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 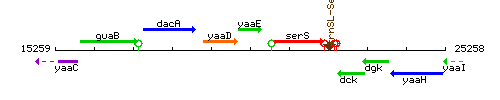
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed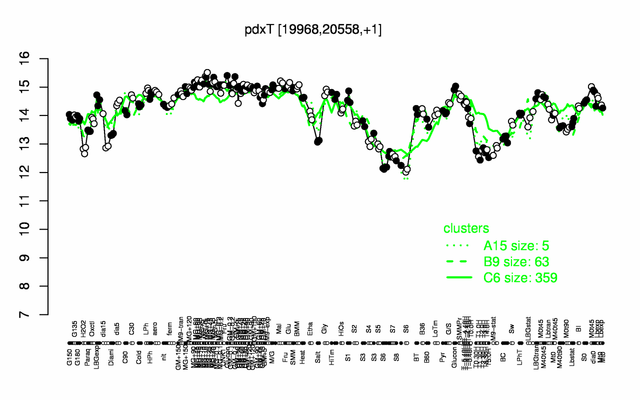
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00120
Phenotypes of a mutant
Database entries
- BsubCyc: BSU00120
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: glutamine amidotransferase pdxT/SNO family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU00120
- UniProt: P37528
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 1683 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 3958 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 749 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 443 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 55 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Miriam Dormeyer, Richard Egelkamp, Martin J Thiele, Elke Hammer, Katrin Gunka, Lorena Stannek, Uwe Völker, Fabian M Commichau
A novel engineering tool in the Bacillus subtilis toolbox: inducer-free activation of gene expression by selection-driven promoter decryptification.
Microbiology (Reading): 2015, 161(Pt 2);354-361
[PubMed:25473090]
[WorldCat.org]
[DOI]
(I p)
Shiori Itagaki, Minami Haga, Yuji Oikawa, Ayaka Sakoda, Yoshie Ohke, Hiroshi Sawada, Tadashi Eguchi, Hideyuki Tamegai
Differences in the roles of a glutamine amidotransferase subunit of pyridoxal 5'-phosphate synthase between Bacillus circulans and Bacillus subtilis.
Biosci Biotechnol Biochem: 2013, 77(7);1481-5
[PubMed:23832367]
[WorldCat.org]
[DOI]
(I p)
Silvia Wallner, Martina Neuwirth, Karlheinz Flicker, Ivo Tews, Peter Macheroux
Dissection of contributions from invariant amino acids to complex formation and catalysis in the heteromeric pyridoxal 5-phosphate synthase complex from Bacillus subtilis.
Biochemistry: 2009, 48(9);1928-35
[PubMed:19152323]
[WorldCat.org]
[DOI]
(I p)
Marco Strohmeier, Thomas Raschle, Jacek Mazurkiewicz, Karsten Rippe, Irmgard Sinning, Teresa B Fitzpatrick, Ivo Tews
Structure of a bacterial pyridoxal 5'-phosphate synthase complex.
Proc Natl Acad Sci U S A: 2006, 103(51);19284-9
[PubMed:17159152]
[WorldCat.org]
[DOI]
(P p)
Thomas Raschle, Nikolaus Amrhein, Teresa B Fitzpatrick
On the two components of pyridoxal 5'-phosphate synthase from Bacillus subtilis.
J Biol Chem: 2005, 280(37);32291-300
[PubMed:16030023]
[WorldCat.org]
[DOI]
(P p)
Jianghai Zhu, John W Burgner, Etti Harms, Boris R Belitsky, Janet L Smith
A new arrangement of (beta/alpha)8 barrels in the synthase subunit of PLP synthase.
J Biol Chem: 2005, 280(30);27914-23
[PubMed:15911615]
[WorldCat.org]
[DOI]
(P p)
Boris R Belitsky
Physical and enzymological interaction of Bacillus subtilis proteins required for de novo pyridoxal 5'-phosphate biosynthesis.
J Bacteriol: 2004, 186(4);1191-6
[PubMed:14762015]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)