PdhD
- Description: dihydrolipoamide dehydrogenase E3 subunit of both pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase complexes
| Gene name | pdhD |
| Synonyms | citL |
| Essential | no |
| Product | dihydrolipoamide dehydrogenase E3 subunit of both pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase complexes |
| Function | links glycolysis and TCA cycle, enzyme in TCA cycle |
| Gene expression levels in SubtiExpress: pdhD | |
| Interactions involving this protein in SubtInteract: PdhD | |
| Metabolic function and regulation of this protein in SubtiPathways: pdhD | |
| MW, pI | 49 kDa, 4.76 |
| Gene length, protein length | 1410 bp, 470 aa |
| Immediate neighbours | pdhC, slp |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 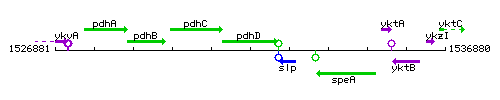
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed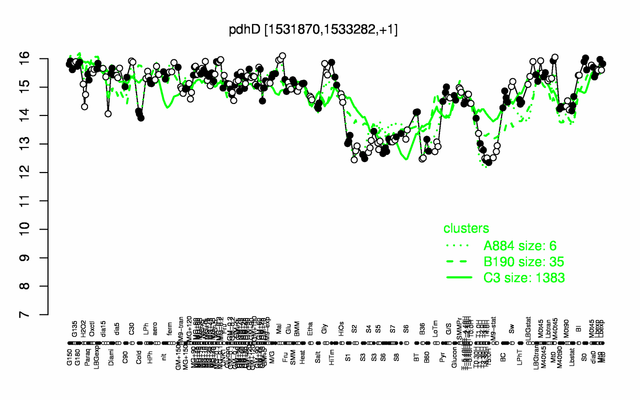
| |
Contents
Categories containing this gene/protein
carbon core metabolism, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14610
Phenotypes of a mutant
- defects in sporulation and unable to grow on glucose as single carbon source PubMed
Database entries
- BsubCyc: BSU14610
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein N(6)-(dihydrolipoyl)lysine + NAD+ = protein N(6)-(lipoyl)lysine + NADH (according to Swiss-Prot)
- Protein family: class-I pyridine nucleotide-disulfide oxidoreductase family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information: Michaelis-Menten PubMed
- Modification: phosphorylated (Ser/Thr/Tyr) PubMed
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU14610
- Structure: 1EBD (complex with binding domain of dihydrolipoamide acetylase, Geobacillus stearothermophilus), 1EBD (complex with binding domain of dihydrolipoamide acetylase, Geobacillus stearothermophilus)
- UniProt: P21880
- KEGG entry: [3]
- E.C. number: 1.8.1.4
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- stringent response: due to presence of guanine at +1 position of the transcript PubMed
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 11483 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 30290 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 11794 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 8779 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 6741 PubMed
Biological materials
- Mutant:
- Expression vector:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Kai Tittmann
Reaction mechanisms of thiamin diphosphate enzymes: redox reactions.
FEBS J: 2009, 276(9);2454-68
[PubMed:19476487]
[WorldCat.org]
[DOI]
(I p)
K F Sheu, J P Blass
The alpha-ketoglutarate dehydrogenase complex.
Ann N Y Acad Sci: 1999, 893;61-78
[PubMed:10672230]
[WorldCat.org]
[DOI]
(P p)
U Neveling, S Bringer-Meyer, H Sahm
Gene and subunit organization of bacterial pyruvate dehydrogenase complexes.
Biochim Biophys Acta: 1998, 1385(2);367-72
[PubMed:9655937]
[WorldCat.org]
[DOI]
(P p)
M S Patel, T E Roche
Molecular biology and biochemistry of pyruvate dehydrogenase complexes.
FASEB J: 1990, 4(14);3224-33
[PubMed:2227213]
[WorldCat.org]
[DOI]
(P p)
P A Frey
Mechanism of coupled electron and group transfer in Escherichia coli pyruvate dehydrogenase.
Ann N Y Acad Sci: 1982, 378;250-64
[PubMed:6805383]
[WorldCat.org]
[DOI]
(P p)
Original publications
Sabine Pérès, Liza Felicori, Franck Molina
Elementary flux modes analysis of functional domain networks allows a better metabolic pathway interpretation.
PLoS One: 2013, 8(10);e76143
[PubMed:24204596]
[WorldCat.org]
[DOI]
(I e)
Frederik M Meyer, Jan Gerwig, Elke Hammer, Christina Herzberg, Fabian M Commichau, Uwe Völker, Jörg Stülke
Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon.
Metab Eng: 2011, 13(1);18-27
[PubMed:20933603]
[WorldCat.org]
[DOI]
(I p)
Shigeo Tojo, Kanako Kumamoto, Kazutake Hirooka, Yasutaro Fujita
Heavy involvement of stringent transcription control depending on the adenine or guanine species of the transcription initiation site in glucose and pyruvate metabolism in Bacillus subtilis.
J Bacteriol: 2010, 192(6);1573-85
[PubMed:20081037]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Haichun Gao, Xin Jiang, Kit Pogliano, Arthur I Aronson
The E1beta and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation.
J Bacteriol: 2002, 184(10);2780-8
[PubMed:11976308]
[WorldCat.org]
[DOI]
(P p)
P N Lowe, J A Hodgson, R N Perham
Dual role of a single multienzyme complex in the oxidative decarboxylation of pyruvate and branched-chain 2-oxo acids in Bacillus subtilis.
Biochem J: 1983, 215(1);133-40
[PubMed:6414463]
[WorldCat.org]
[DOI]
(P p)