Difference between revisions of "PdhC"
(→Database entries) |
(→Database entries) |
||
| Line 78: | Line 78: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1W88 1W88] (E1 in complex with subunit binding domain of E2, ''Geobacillus stearothermophilus''), [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId 2PDE] (peripheral subunit binding domain, ''Geobacillus stearothermophilus''), [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId 1LAC] (lipoyl domain, ''Geobacillus stearothermophilus'') | + | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1W88 1W88] (E1 in complex with subunit binding domain of E2, ''Geobacillus stearothermophilus''), [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId 2PDE] (peripheral subunit binding domain, ''Geobacillus stearothermophilus''), [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId 1LAC] (lipoyl domain, ''Geobacillus stearothermophilus''), [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId 1B5S] (catalytic domain (residues 184-425) , ''Geobacillus stearothermophilus'') |
* '''Swiss prot entry:''' | * '''Swiss prot entry:''' | ||
Revision as of 15:22, 9 February 2009
- Description: pyruvate dehydrogenase (dihydrolipoamide acetyltransferase E2 subunit)
| Gene name | pdhC |
| Synonyms | |
| Essential | no |
| Product | pyruvate dehydrogenase (dihydrolipoamide acetyltransferase E2 subunit) |
| Function | links glycolysis and TCA cycle |
| MW, pI | 47 kDa, 4.855 |
| Gene length, protein length | 1326 bp, 442 aa |
| Immediate neighbours | pdhB, pdhD |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 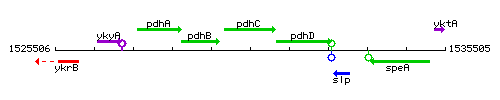
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
defects in sporulation and unable to grow on glucose as single carbon source PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane associated PubMed
Database entries
- Structure: 1W88 (E1 in complex with subunit binding domain of E2, Geobacillus stearothermophilus), 2PDE (peripheral subunit binding domain, Geobacillus stearothermophilus), 1LAC (lipoyl domain, Geobacillus stearothermophilus), 1B5S (catalytic domain (residues 184-425) , Geobacillus stearothermophilus)
- Swiss prot entry:
- KEGG entry:
- E.C. number: 2.3.1.12
Additional information
Expression and regulation
- Sigma factor: SigA
- Regulation: weak induction by glucose PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6: 697-707 PubMed
- Hahne et al. (2008) From complementarity to comprehensiveness - targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics 8: 4123-4136 PubMed
- Gao et al. (2002) The E1beta and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation.J. Bacteriol. 184: 2780-2788. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed