Difference between revisions of "PdhB"
(→Basic information/ Evolution) |
(→Basic information/ Evolution) |
||
| Line 81: | Line 81: | ||
* '''Protein family:''' | * '''Protein family:''' | ||
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' [[AcoB]], [[BkdAB]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
Revision as of 19:08, 8 June 2013
- Description: pyruvate dehydrogenase (E1 beta subunit)
| Gene name | pdhB |
| Synonyms | |
| Essential | no |
| Product | pyruvate dehydrogenase (E1 beta subunit) |
| Function | links glycolysis and TCA cycle |
| Gene expression levels in SubtiExpress: pdhB | |
| Interactions involving this protein in SubtInteract: PdhB | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 35 kDa, 4.547 |
| Gene length, protein length | 975 bp, 325 aa |
| Immediate neighbours | pdhA, pdhC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 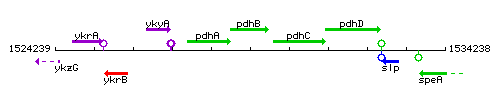
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14590
Phenotypes of a mutant
- defects in sporulation and unable to grow on glucose as single carbon source PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Pyruvate + [dihydrolipoyllysine-residue acetyltransferase] lipoyllysine = [dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine + CO2 (according to Swiss-Prot)
- Protein family:
Extended information on the protein
- Kinetic information: Michaelis-Menten PubMed
- Domains:
- Modification: phosphorylation on (Ser-302 OR Ser-306) PubMed
- Cofactor(s):
- Effectors of protein activity:
- Inhibited thiamine 2-thiothiazolone diphosphate and NADH PubMed
- Low sensibility to NADPH
- Localization: membrane associated PubMed
Database entries
- Structure: 1W88 (E1 in complex with subunit binding domain of E2, Geobacillus stearothermophilus)
- UniProt: P21882
- KEGG entry: [3]
- E.C. number: 1.2.4.1
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- stringent response: due to presence of guanine at +1 position of the transcript PubMed
- Additional information:
Biological materials
- Mutant: GP459 (spc), available in Stülke lab
- Expression vector:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Arthur Aronson, Purdue University, West Lafayette, USA homepage
Your additional remarks
References
Reviews
Original publications
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Haichun Gao, Xin Jiang, Kit Pogliano, Arthur I Aronson
The E1beta and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation.
J Bacteriol: 2002, 184(10);2780-8
[PubMed:11976308]
[WorldCat.org]
[DOI]
(P p)
M M Nakano, Y P Dailly, P Zuber, D P Clark
Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth.
J Bacteriol: 1997, 179(21);6749-55
[PubMed:9352926]
[WorldCat.org]
[DOI]
(P p)
P N Lowe, J A Hodgson, R N Perham
Dual role of a single multienzyme complex in the oxidative decarboxylation of pyruvate and branched-chain 2-oxo acids in Bacillus subtilis.
Biochem J: 1983, 215(1);133-40
[PubMed:6414463]
[WorldCat.org]
[DOI]
(P p)