Difference between revisions of "PdaA"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 42: | Line 42: | ||
* '''Locus tag:''' BSU07980 | * '''Locus tag:''' BSU07980 | ||
| − | |||
| − | |||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
Revision as of 20:29, 27 January 2012
- Description: polysaccharide deacetylase, required for germination
| Gene name | pdaA |
| Synonyms | yfjS |
| Essential | no |
| Product | polysaccharide deacetylase |
| Function | germination |
| MW, pI | 29 kDa, 7.318 |
| Gene length, protein length | 789 bp, 263 aa |
| Immediate neighbours | yfjT, yfjR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 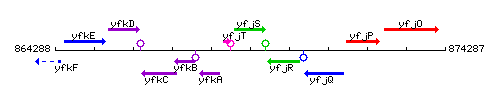
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU07980
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: polysaccharide deacetylase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 1W17
- UniProt: O34928
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: pdaA PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Leif Steil, Mónica Serrano, Adriano O Henriques, Uwe Völker
Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 2);399-420
[PubMed:15699190]
[WorldCat.org]
[DOI]
(P p)
Tatsuya Fukushima, Toshihiko Kitajima, Junichi Sekiguchi
A polysaccharide deacetylase homologue, PdaA, in Bacillus subtilis acts as an N-acetylmuramic acid deacetylase in vitro.
J Bacteriol: 2005, 187(4);1287-92
[PubMed:15687192]
[WorldCat.org]
[DOI]
(P p)
David E Blair, Daan M F van Aalten
Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine.
FEBS Lett: 2004, 570(1-3);13-9
[PubMed:15251431]
[WorldCat.org]
[DOI]
(P p)
Meghan E Gilmore, Dabanjan Bandyopadhyay, Amanda M Dean, Sarah D Linnstaedt, David L Popham
Production of muramic delta-lactam in Bacillus subtilis spore peptidoglycan.
J Bacteriol: 2004, 186(1);80-9
[PubMed:14679227]
[WorldCat.org]
[DOI]
(P p)
Tatsuya Fukushima, Hiroki Yamamoto, Abdelmadjid Atrih, Simon J Foster, Junichi Sekiguchi
A polysaccharide deacetylase gene (pdaA) is required for germination and for production of muramic delta-lactam residues in the spore cortex of Bacillus subtilis.
J Bacteriol: 2002, 184(21);6007-15
[PubMed:12374835]
[WorldCat.org]
[DOI]
(P p)