Difference between revisions of "PbuG"
| Line 23: | Line 23: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[guaA]]'', ''[[yebC]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[guaA]]'', ''[[yebC]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU06370 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU06370 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU06370 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU06370 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU06370 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU06370 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:pbuG_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:pbuG_context.gif]] | ||
Revision as of 09:37, 14 May 2013
- Description: hypoxanthin/ guanine permease
| Gene name | pbuG |
| Synonyms | yebB |
| Essential | no |
| Product | hypoxanthin/ guanine permease |
| Function | hypoxanthine and guanine uptake |
| Gene expression levels in SubtiExpress: pbuG | |
| Metabolic function and regulation of this protein in SubtiPathways: Purine salvage, Nucleotides (regulation) | |
| MW, pI | 46 kDa, 8.808 |
| Gene length, protein length | 1320 bp, 440 aa |
| Immediate neighbours | guaA, yebC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed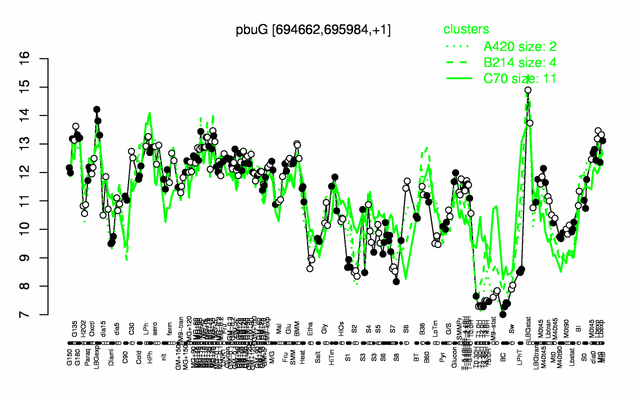
| |
Contents
Categories containing this gene/protein
transporters/ other, biosynthesis/ acquisition of nucleotides, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU06370
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): PbuO
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane PubMed
Database entries
- Structure: 2EEW (the pbuG riboswitch, U47c mutant, bound to hypoxanthine)
- UniProt: O34987
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: pbuG PubMed
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- PurR: transcription repression (molecular inducer: PRPP) PubMed
- G-box: transcription termination/ antitermination (riboswitch) PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Lars Engholm Johansen, Per Nygaard, Catharina Lassen, Yvonne Agersø, Hans H Saxild
Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL).
J Bacteriol: 2003, 185(17);5200-9
[PubMed:12923093]
[WorldCat.org]
[DOI]
(P p)
Maumita Mandal, Benjamin Boese, Jeffrey E Barrick, Wade C Winkler, Ronald R Breaker
Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria.
Cell: 2003, 113(5);577-86
[PubMed:12787499]
[WorldCat.org]
[DOI]
(P p)
H H Saxild, K Brunstedt, K I Nielsen, H Jarmer, P Nygaard
Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO.
J Bacteriol: 2001, 183(21);6175-83
[PubMed:11591660]
[WorldCat.org]
[DOI]
(P p)