Difference between revisions of "PbpA"
| Line 1: | Line 1: | ||
| − | * '''Description:''' class B [[penicillin-binding protein]] PBP 2A <br/><br/> | + | * '''Description:''' class B [[penicillin-binding protein]] PBP 2A, required for [[cell wall synthesis]] during cell elongation <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 67: | Line 67: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| + | ** required for [[cell wall synthesis]] during cell elongation {{PubMed|12896990}} | ||
| + | ** driver of [[MreB]] dynamics {{PubMed|21636744,21636745}} | ||
* '''Protein family:''' | * '''Protein family:''' | ||
| Line 85: | Line 87: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| − | ** part of the [[ | + | ** part of the [[elongasome]] {{PubMed|21636744,21636745}} |
** folding requires [[PrsA]] {{PubMed|20487272}} | ** folding requires [[PrsA]] {{PubMed|20487272}} | ||
| − | * '''[[Localization]]:''' extracellular (signal peptide) [http://www.ncbi.nlm.nih.gov/pubmed/18957862 PubMed] | + | * '''[[Localization]]:''' |
| + | ** extracellular (signal peptide) [http://www.ncbi.nlm.nih.gov/pubmed/18957862 PubMed] | ||
| + | ** localization depends on the availability of peptidoglyan precursors (lipid II) {{PubMed|23895585}} | ||
=== Database entries === | === Database entries === | ||
Revision as of 09:24, 5 March 2014
- Description: class B penicillin-binding protein PBP 2A, required for cell wall synthesis during cell elongation
| Gene name | pbpA |
| Synonyms | yqgF |
| Essential | no |
| Product | penicillin-binding protein PBP 2A |
| Function | formation of a rod-shaped peptidoglycan cell wall, spore outgrowth |
| Gene expression levels in SubtiExpress: pbpA | |
| Interactions involving this protein in SubtInteract: PbpA | |
| MW, pI | 79 kDa, 9.571 |
| Gene length, protein length | 2148 bp, 716 aa |
| Immediate neighbours | pstS, yqgE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 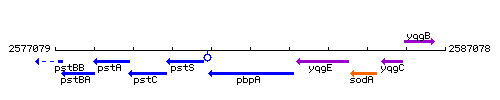
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25000
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- required for cell wall synthesis during cell elongation PubMed
- driver of MreB dynamics PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Interactions:
- part of the elongasome PubMed
- folding requires PrsA PubMed
Database entries
- Structure:
- UniProt: P54488
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jeff Errington lab
- Antibody:
Labs working on this gene/protein
Jeff Errington, Newcastle University, UK homepage
Your additional remarks
References
Marta Carolina Afonso Lages, Katrin Beilharz, Danae Morales Angeles, Jan-Willem Veening, Dirk-Jan Scheffers
The localization of key Bacillus subtilis penicillin binding proteins during cell growth is determined by substrate availability.
Environ Microbiol: 2013, 15(12);3272-81
[PubMed:23895585]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Ethan C Garner, Remi Bernard, Wenqin Wang, Xiaowei Zhuang, David Z Rudner, Tim Mitchison
Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis.
Science: 2011, 333(6039);222-5
[PubMed:21636745]
[WorldCat.org]
[DOI]
(I p)
Julia Domínguez-Escobar, Arnaud Chastanet, Alvaro H Crevenna, Vincent Fromion, Roland Wedlich-Söldner, Rut Carballido-López
Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria.
Science: 2011, 333(6039);225-8
[PubMed:21636744]
[WorldCat.org]
[DOI]
(I p)
Hanne-Leena Hyyryläinen, Bogumila C Marciniak, Kathleen Dahncke, Milla Pietiäinen, Pascal Courtin, Marika Vitikainen, Raili Seppala, Andreas Otto, Dörte Becher, Marie-Pierre Chapot-Chartier, Oscar P Kuipers, Vesa P Kontinen
Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis.
Mol Microbiol: 2010, 77(1);108-27
[PubMed:20487272]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Yuping Wei, Teresa Havasy, Derrell C McPherson, David L Popham
Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbpA and pbpH.
J Bacteriol: 2003, 185(16);4717-26
[PubMed:12896990]
[WorldCat.org]
[DOI]
(P p)
T Murray, D L Popham, C B Pearson, A R Hand, P Setlow
Analysis of outgrowth of Bacillus subtilis spores lacking penicillin-binding protein 2a.
J Bacteriol: 1998, 180(24);6493-502
[PubMed:9851991]
[WorldCat.org]
[DOI]
(P p)
T Murray, D L Popham, P Setlow
Identification and characterization of pbpA encoding Bacillus subtilis penicillin-binding protein 2A.
J Bacteriol: 1997, 179(9);3021-9
[PubMed:9139922]
[WorldCat.org]
[DOI]
(P p)