Difference between revisions of "PBQ200"
| Line 6: | Line 6: | ||
Sequencing primers: | Sequencing primers: | ||
| − | *'''M13_puc_for:''' 5‘-GTAAAACGACGGCCAGTG-3‘ | + | *'''M13_puc_for:''' 5‘-GTAAAACGACGGCCAGTG-3‘ |
| − | *'''M13_puc_rev:''' 5‘-GGAAACAGCTATGACCATG-3‘ | + | *'''M13_puc_rev:''' 5‘-GGAAACAGCTATGACCATG-3‘ |
| + | *'''FX125 (rev; primes -55bp of MCS):''' 5‘-GGCTCGTATGTTGTGTGG-3‘ | ||
'''Reference:''' | '''Reference:''' | ||
<pubmed> 8057358 </pubmed> | <pubmed> 8057358 </pubmed> | ||
Revision as of 17:17, 21 August 2012
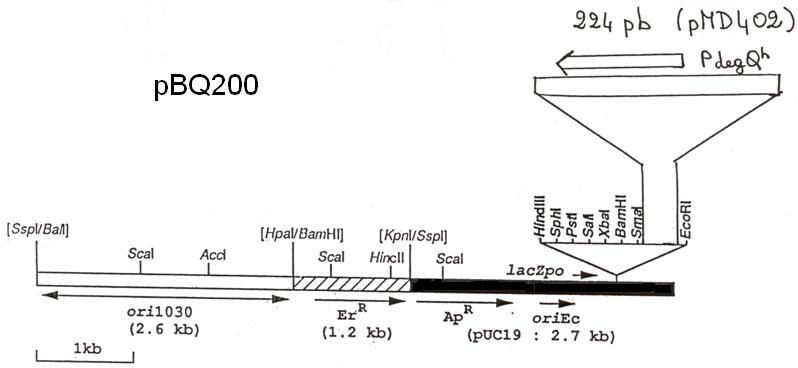
Application:
The vector was constructed in the George Rapoport lab and it is suitable for constitutive overexpression of proteins in B. subtilis. The plasmid confers resistance to ampicillin and erythromycin in E. coli and B. subtilis, respectively. pBQ200 is based on the vector pHT315. A Shine-Dalgarno sequence has to be fused to the open reading frame during PCR.
Sequencing primers:
- M13_puc_for: 5‘-GTAAAACGACGGCCAGTG-3‘
- M13_puc_rev: 5‘-GGAAACAGCTATGACCATG-3‘
- FX125 (rev; primes -55bp of MCS): 5‘-GGCTCGTATGTTGTGTGG-3‘
Reference: