Difference between revisions of "NasF"
(→Extended information on the protein) |
(→References) |
||
| Line 139: | Line 139: | ||
=References= | =References= | ||
| − | + | ==Reviews== | |
| + | <pubmed> 22103536 </pubmed> | ||
| + | ==Original publications== | ||
<pubmed>10217486,12823818,,8799114,9765565,10972836,16885456,7868621 21091510 </pubmed> | <pubmed>10217486,12823818,,8799114,9765565,10972836,16885456,7868621 21091510 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:43, 23 November 2011
- Description: uroporphyrinogen methyltransferase
| Gene name | nasF |
| Synonyms | nasBE |
| Essential | no |
| Product | uroporphyrinogen methyltransferase |
| Function | nitrate respiration |
| Metabolic function and regulation of this protein in SubtiPathways: Alternative nitrogen sources | |
| MW, pI | 53 kDa, 6.523 |
| Gene length, protein length | 1449 bp, 483 aa |
| Immediate neighbours | ycgT, nasE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 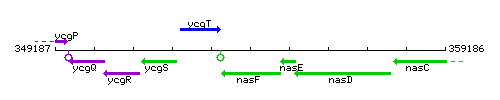
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
respiration, utilization of nitrogen sources other than amino acids
This gene is a member of the following regulons
Fur regulon, NsrR regulon, ResD regulon, TnrA regulon
The gene
Basic information
- Locus tag: BSU03280
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: S-adenosyl-L-methionine + uroporphyrinogen III = S-adenosyl-L-homocysteine + precorrin-1 (according to Swiss-Prot)
- Protein family: precorrin methyltransferase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P42437
- KEGG entry: [3]
- E.C. number: 2.1.1.107
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Víctor M Luque-Almagro, Andrew J Gates, Conrado Moreno-Vivián, Stuart J Ferguson, David J Richardson, M Dolores Roldán
Bacterial nitrate assimilation: gene distribution and regulation.
Biochem Soc Trans: 2011, 39(6);1838-43
[PubMed:22103536]
[WorldCat.org]
[DOI]
(I p)
Original publications
Sushma Kommineni, Erik Yukl, Takahiro Hayashi, Jacob Delepine, Hao Geng, Pierre Moënne-Loccoz, Michiko M Nakano
Nitric oxide-sensitive and -insensitive interaction of Bacillus subtilis NsrR with a ResDE-controlled promoter.
Mol Microbiol: 2010, 78(5);1280-93
[PubMed:21091510]
[WorldCat.org]
[DOI]
(I p)
Michiko M Nakano, Hao Geng, Shunji Nakano, Kazuo Kobayashi
The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression.
J Bacteriol: 2006, 188(16);5878-87
[PubMed:16885456]
[WorldCat.org]
[DOI]
(P p)
Ken-ichi Yoshida, Hirotake Yamaguchi, Masaki Kinehara, Yo-hei Ohki, Yoshiko Nakaura, Yasutaro Fujita
Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box.
Mol Microbiol: 2003, 49(1);157-65
[PubMed:12823818]
[WorldCat.org]
[DOI]
(P p)
M M Nakano, Y Zhu, M Lacelle, X Zhang, F M Hulett
Interaction of ResD with regulatory regions of anaerobically induced genes in Bacillus subtilis.
Mol Microbiol: 2000, 37(5);1198-207
[PubMed:10972836]
[WorldCat.org]
[DOI]
(P p)
Per Johansson, Lars Hederstedt
Organization of genes for tetrapyrrole biosynthesis in gram--positive bacteria.
Microbiology (Reading): 1999, 145 ( Pt 3);529-538
[PubMed:10217486]
[WorldCat.org]
[DOI]
(P p)
M M Nakano, T Hoffmann, Y Zhu, D Jahn
Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE.
J Bacteriol: 1998, 180(20);5344-50
[PubMed:9765565]
[WorldCat.org]
[DOI]
(P p)
L V Wray, A E Ferson, K Rohrer, S H Fisher
TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1996, 93(17);8841-5
[PubMed:8799114]
[WorldCat.org]
[DOI]
(P p)
K Ogawa, E Akagawa, K Yamane, Z W Sun, M LaCelle, P Zuber, M M Nakano
The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis.
J Bacteriol: 1995, 177(5);1409-13
[PubMed:7868621]
[WorldCat.org]
[DOI]
(P p)