MutS
- Description: DNA mismatch repair (mismatch recognition protein)
| Gene name | mutS |
| Synonyms | |
| Essential | no |
| Product | mismatch recognition protein |
| Function | DNA repair |
| Gene expression levels in SubtiExpress: mutS | |
| Interactions involving this protein in SubtInteract: MutS | |
| MW, pI | 97 kDa, 5.189 |
| Gene length, protein length | 2574 bp, 858 aa |
| Immediate neighbours | cotE, mutL |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 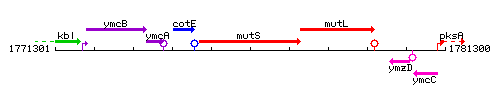
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed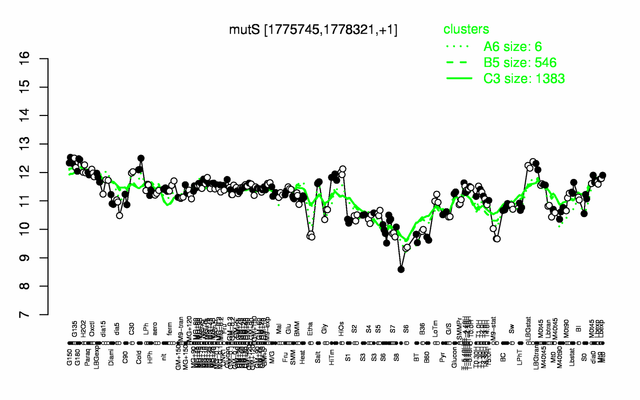
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU17040
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- MutS recognizes mismatches in the DNA and recruits MutL to the site of mismatch recognition
- Protein family: DNA mismatch repair mutS family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P49849
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- An antisense RNA is predicted for mutS PubMed
Biological materials
- Mutant: GP1190 (del mutSL::aphA3) available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Justin S Lenhart, Anushi Sharma, Manju M Hingorani, Lyle A Simmons
DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication.
Mol Microbiol: 2013, 87(3);553-68
[PubMed:23228104]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Nicole M Dupes, Brian W Walsh, Andrew D Klocko, Justin S Lenhart, Heather L Peterson, David A Gessert, Cassie E Pavlick, Lyle A Simmons
Mutations in the Bacillus subtilis beta clamp that separate its roles in DNA replication from mismatch repair.
J Bacteriol: 2010, 192(13);3452-63
[PubMed:20453097]
[WorldCat.org]
[DOI]
(I p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Mario Pedraza-Reyes, Ronald E Yasbin
Contribution of the mismatch DNA repair system to the generation of stationary-phase-induced mutants of Bacillus subtilis.
J Bacteriol: 2004, 186(19);6485-91
[PubMed:15375129]
[WorldCat.org]
[DOI]
(P p)
F Ginetti, M Perego, A M Albertini, A Galizzi
Bacillus subtilis mutS mutL operon: identification, nucleotide sequence and mutagenesis.
Microbiology (Reading): 1996, 142 ( Pt 8);2021-9
[PubMed:8760914]
[WorldCat.org]
[DOI]
(P p)