Difference between revisions of "MurJ"
| Line 59: | Line 59: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU30050&redirect=T BSU30050] | ||
* '''DBTBS entry:''' no entry | * '''DBTBS entry:''' no entry | ||
| Line 96: | Line 97: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU30050&redirect=T BSU30050] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 14:32, 2 April 2014
- Description: similar to spore cortex protein
| Gene name | ytgP |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| Gene expression levels in SubtiExpress: ytgP | |
| MW, pI | 59 kDa, 10.089 |
| Gene length, protein length | 1632 bp, 544 aa |
| Immediate neighbours | ytzG, ytfP |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 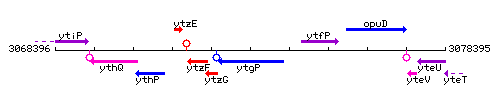
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU30050
Phenotypes of a mutant
Mutants lacking YtgP produce long cells and chains of cells, suggesting a role in cell division. PubMed
Database entries
- BsubCyc: BSU30050
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU30050
- Structure:
- UniProt: O34674
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Allison Fay, Jonathan Dworkin
Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth.
J Bacteriol: 2009, 191(19);6020-8
[PubMed:19666716]
[WorldCat.org]
[DOI]
(I p)
Pradeep Vasudevan, Jessica McElligott, Christa Attkisson, Michael Betteken, David L Popham
Homologues of the Bacillus subtilis SpoVB protein are involved in cell wall metabolism.
J Bacteriol: 2009, 191(19);6012-9
[PubMed:19648239]
[WorldCat.org]
[DOI]
(I p)