Difference between revisions of "MtlD"
| Line 82: | Line 82: | ||
* '''Swiss prot entry:''' | * '''Swiss prot entry:''' | ||
| − | * '''KEGG entry:''' | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU03990] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
Revision as of 10:22, 20 March 2009
- Description: mannitol-1-phosphate 5-dehydrogenase
| Gene name | mtlD |
| Synonyms | mtlB |
| Essential | no |
| Product | mannitol-1-phosphate 5-dehydrogenase |
| Function | mannitol utilization |
| MW, pI | 40 kDa, 6.291 |
| Gene length, protein length | 1098 bp, 366 aa |
| Immediate neighbours | mtlA, ycsA |
| Gene sequence (+200bp) | Protein sequence |
| Caution: The sequence for this gene in SubtiList contains errors | |
Genetic context 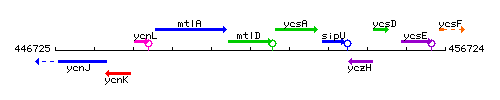
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Watanabe S, Hamano M, Kakeshita H, Bunai K, Tojo S, Yamaguchi H, Fujita Y, Wong SL, Yamane K. (2003) Mannitol-1-phosphate dehydrogenase (MtlD) is required for mannitol and glucitol assimilation in Bacillus subtilis: possible cooperation of mtl and gut operons. J Bacteriol. Aug;185(16): 4816-24. PubMed