Difference between revisions of "MtlD"
| Line 56: | Line 56: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 110: | Line 107: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
** [[MtlR]]: transcription activation {{PubMed|20444094}} | ** [[MtlR]]: transcription activation {{PubMed|20444094}} | ||
| + | |||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** An [[ncRNA|antisense RNA]] is predicted for ''[[mtlD]]'' {{PubMed|20525796}} | ||
=Biological materials = | =Biological materials = | ||
| Line 132: | Line 131: | ||
=References= | =References= | ||
| − | <pubmed>12897001, 20444094,10627040 </pubmed> | + | <pubmed>12897001, 20444094,10627040 20525796</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:39, 4 May 2011
- Description: mannitol-1-phosphate 5-dehydrogenase
| Gene name | mtlD |
| Synonyms | mtlB |
| Essential | no |
| Product | mannitol-1-phosphate 5-dehydrogenase |
| Function | mannitol utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 40 kDa, 6.291 |
| Gene length, protein length | 1098 bp, 366 aa |
| Immediate neighbours | mtlF, ycsA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 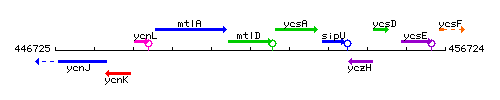
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU03990
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: D-mannitol 1-phosphate + NAD+ = D-fructose 6-phosphate + NADH (according to Swiss-Prot)
- Protein family: mannitol dehydrogenase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure: 3H2Z (from Shigella flexneri 2a str. 2457t mutant, 42% identity, 59% similarity)
- UniProt: P42957
- KEGG entry: [3]
- E.C. number: 1.1.1.17
Additional information
Expression and regulation
- Additional information:
- An antisense RNA is predicted for mtlD PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Philippe Joyet, Meriem Derkaoui, Sandrine Poncet, Josef Deutscher
Control of Bacillus subtilis mtl operon expression by complex phosphorylation-dependent regulation of the transcriptional activator MtlR.
Mol Microbiol: 2010, 76(5);1279-94
[PubMed:20444094]
[WorldCat.org]
[DOI]
(I p)
Shouji Watanabe, Miyuki Hamano, Hiroshi Kakeshita, Keigo Bunai, Shigeo Tojo, Hirotake Yamaguchi, Yasutaro Fujita, Sui-Lam Wong, Kunio Yamane
Mannitol-1-phosphate dehydrogenase (MtlD) is required for mannitol and glucitol assimilation in Bacillus subtilis: possible cooperation of mtl and gut operons.
J Bacteriol: 2003, 185(16);4816-24
[PubMed:12897001]
[WorldCat.org]
[DOI]
(P p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)