Difference between revisions of "MrnC"
| Line 59: | Line 59: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU00950&redirect=T BSU00950] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gltX-cysES-yazC-yacOP.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gltX-cysES-yazC-yacOP.html] | ||
| Line 96: | Line 97: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU00950&redirect=T BSU00950] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 12:48, 2 April 2014
- Description: RNase Mini-III
| Gene name | mrnC |
| Synonyms | yazC |
| Essential | no |
| Product | RNase Mini-III |
| Function | maturation of 23S rRNA |
| Gene expression levels in SubtiExpress: mrnC | |
| Metabolic function and regulation of this protein in SubtiPathways: mrnC | |
| MW, pI | 16 kDa, 9.153 |
| Gene length, protein length | 429 bp, 143 aa |
| Immediate neighbours | cysS, yacO |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed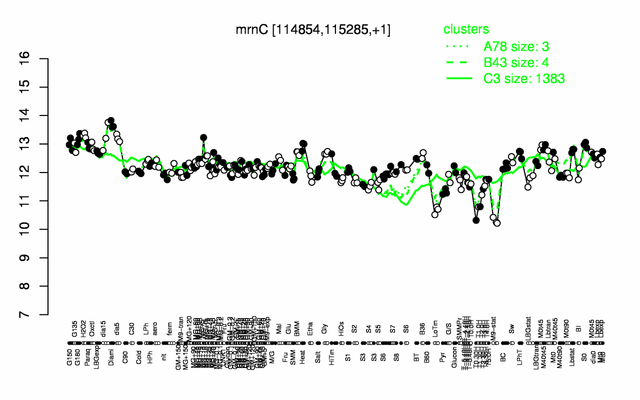
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00950
Phenotypes of a mutant
Database entries
- BsubCyc: BSU00950
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: RNase III family (but lacks the dsRNA-binding domain)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: RplC binding to the precursor 23S rRNA stimulates MrnC activity PubMed
Database entries
- BsubCyc: BSU00950
- Structure:
- UniProt: O31418
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism: T-box, overlaps a transcription terminator upstream of cysE: transcription antitermination PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Ciaran Condon, IBPC, Paris, France Homepage
David Bechhofer, Mount Sinai School, New York, USA Homepage
Your additional remarks
References
Yulia Redko, Ciarán Condon
Maturation of 23S rRNA in Bacillus subtilis in the absence of Mini-III.
J Bacteriol: 2010, 192(1);356-9
[PubMed:19880604]
[WorldCat.org]
[DOI]
(I p)
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)
Yulia Redko, Ciarán Condon
Ribosomal protein L3 bound to 23S precursor rRNA stimulates its maturation by Mini-III ribonuclease.
Mol Microbiol: 2009, 71(5);1145-54
[PubMed:19154332]
[WorldCat.org]
[DOI]
(I p)
Gabriela Olmedo, Plinio Guzmán
Mini-III, a fourth class of RNase III catalyses maturation of the Bacillus subtilis 23S ribosomal RNA.
Mol Microbiol: 2008, 68(5);1073-6
[PubMed:18430137]
[WorldCat.org]
[DOI]
(I p)
Yulia Redko, David H Bechhofer, Ciarán Condon
Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis.
Mol Microbiol: 2008, 68(5);1096-106
[PubMed:18363798]
[WorldCat.org]
[DOI]
(I p)
L Chen, L P James, J D Helmann
Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions.
J Bacteriol: 1993, 175(17);5428-37
[PubMed:8396117]
[WorldCat.org]
[DOI]
(P p)