Difference between revisions of "MreD"
(→Labs working on this gene/protein) |
|||
| Line 126: | Line 126: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | |||
| + | [[Peter Graumann]], Freiburg University, Germany [http://www.biologie.uni-freiburg.de/data/bio2/graumann/index.htm homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
Revision as of 08:19, 8 April 2009
- Description: MreD is a cell shape determining protein and is associated with the MreB cytoskeleton in B. subtilis and other rod shaped bacteria.
| Gene name | mreD |
| Synonyms | rodB |
| Essential | yes |
| Product | cell-shape determining protein |
| Function | cell-shape determation |
| MW, pI | 19 kDa, 7.906 |
| Gene length, protein length | 516 bp, 172 aa |
| Immediate neighbours | minC, mreC |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 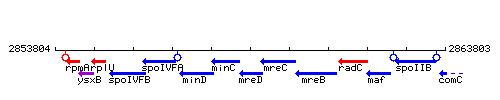
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates: 2858547-2859062
Phenotypes of a mutant
the phenotype of mreD is similar to that of mreC. mreD is essential under normal conditions PubMed. Depletion of MreD leads to a progressive increase in the width and a decrease in the length of the cell and cells become lytic. In the depletion strain, lysis can be prevented and cell growth, but not cell shape, can be recovered by inculaion of Magnesium in the media. This shape defect is consistent with a role for mreD in cell wall synthesis during elongation and has a similar phenotype to other genes with roles in elongation.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
Function
MreD functions in cell wall synthesis by, together with the MreB cytoskeleton, localizing the cell wall synthetic machinery to the correct part of the cell. As a transmembrane protein MreD is thought to provide a patch on the membrane that MreB inteacts with. MreC therefore ensures that the cell wall is made in the correct way to maintain the proper shape of the cell.
mreD and the mapping of mre-min operon
The mre-min operon was first sequenced by levin et al in 1992. The temperature sentitive rodB1 mutation mapperd to that region.
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: None/ structural
- Protein family: COG2891
- Paralogous protein(s): None
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure: None
- Swiss prot entry: Q01467
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: A conditional mutant with an inframe deletion of mreD complemented by a xylose inducible copy at an ectopic locus (strain named 4352) is avaliable from the Errington Group.
- Expression vector:
- lacZ fusion:
- GFP fusion: A functional N-terminal GFP fusion has been made where the fusion protein is the only copy of the gene in the cell: strain 3416 PubMed.
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Peter Graumann, Freiburg University, Germany homepage
Your additional remarks
References
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed