Difference between revisions of "MifM"
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || sensor of [[SpoIIIJ]] activity | |style="background:#ABCDEF;" align="center"| '''Product''' || sensor of [[SpoIIIJ]] activity | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || control of ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Function''' || control of ''[[yidC2]]'' translation |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 11 kDa, 5.829 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 11 kDa, 5.829 | ||
| Line 84: | Line 84: | ||
** the C-terminus of MifM interacts with the ribosomal polypeptide exit tunnel {{PubMed|19779460}} | ** the C-terminus of MifM interacts with the ribosomal polypeptide exit tunnel {{PubMed|19779460}} | ||
| − | * '''[[Localization]]:''' membrane {{PubMed|19779460}} | + | * '''[[Localization]]:''' |
| + | ** membrane {{PubMed|19779460}} | ||
=== Database entries === | === Database entries === | ||
Revision as of 10:37, 7 August 2012
- Description: ribosome-nascent chain sensor of membrane protein biogenesis
| Gene name | mifM |
| Synonyms | yqzJ |
| Essential | no |
| Product | sensor of SpoIIIJ activity |
| Function | control of yidC2 translation |
| MW, pI | 11 kDa, 5.829 |
| Gene length, protein length | 285 bp, 95 aa |
| Immediate neighbours | polY1, yidC2 |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 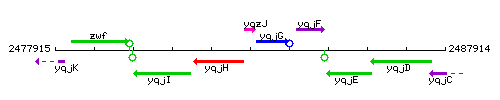
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
translation, membrane proteins
This gene is a member of the following regulons
The MifM regulon: yqjG
The gene
Basic information
- Locus tag: BSU23880
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: acts as a kind of "leader peptide" that does or does not allow translation through a hairpin structure in the yqjG mRNA PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- the C-terminus of MifM interacts with the ribosomal polypeptide exit tunnel PubMed
- Localization:
- membrane PubMed
Database entries
- Structure:
- UniProt: Q7WY64
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- An antisense RNA is predicted for mifM PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Shinobu Chiba, Koreaki Ito
Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM.
Mol Cell: 2012, 47(6);863-72
[PubMed:22864117]
[WorldCat.org]
[DOI]
(I p)
Gene-Wei Li, Eugene Oh, Jonathan S Weissman
The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria.
Nature: 2012, 484(7395);538-41
[PubMed:22456704]
[WorldCat.org]
[DOI]
(I e)
Pierre Nicolas, Ulrike Mäder, Etienne Dervyn, Tatiana Rochat, Aurélie Leduc, Nathalie Pigeonneau, Elena Bidnenko, Elodie Marchadier, Mark Hoebeke, Stéphane Aymerich, Dörte Becher, Paola Bisicchia, Eric Botella, Olivier Delumeau, Geoff Doherty, Emma L Denham, Mark J Fogg, Vincent Fromion, Anne Goelzer, Annette Hansen, Elisabeth Härtig, Colin R Harwood, Georg Homuth, Hanne Jarmer, Matthieu Jules, Edda Klipp, Ludovic Le Chat, François Lecointe, Peter Lewis, Wolfram Liebermeister, Anika March, Ruben A T Mars, Priyanka Nannapaneni, David Noone, Susanne Pohl, Bernd Rinn, Frank Rügheimer, Praveen K Sappa, Franck Samson, Marc Schaffer, Benno Schwikowski, Leif Steil, Jörg Stülke, Thomas Wiegert, Kevin M Devine, Anthony J Wilkinson, Jan Maarten van Dijl, Michael Hecker, Uwe Völker, Philippe Bessières, Philippe Noirot
Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis.
Science: 2012, 335(6072);1103-6
[PubMed:22383849]
[WorldCat.org]
[DOI]
(I p)
Shinobu Chiba, Takashi Kanamori, Takuya Ueda, Yoshinori Akiyama, Kit Pogliano, Koreaki Ito
Recruitment of a species-specific translational arrest module to monitor different cellular processes.
Proc Natl Acad Sci U S A: 2011, 108(15);6073-8
[PubMed:21383133]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Shinobu Chiba, Anne Lamsa, Kit Pogliano
A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis.
EMBO J: 2009, 28(22);3461-75
[PubMed:19779460]
[WorldCat.org]
[DOI]
(I p)