MgsA
- Description: methylglyoxal synthase
| Gene name | mgsA |
| Synonyms | ypjF, jojF |
| Essential | no |
| Product | methylglyoxal synthase |
| Function | bypass of glycolysis |
| Gene expression levels in SubtiExpress: mgsA | |
| Interactions involving this protein in SubtInteract: MgsA | |
| Metabolic function and regulation of this protein in SubtiPathways: mgsA | |
| MW, pI | 14 kDa, 4.919 |
| Gene length, protein length | 411 bp, 137 aa |
| Immediate neighbours | bshB1, dapB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 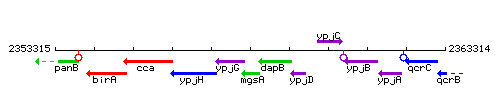
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22480
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Glycerone phosphate = methylglyoxal + phosphate (according to Swiss-Prot)
- Protein family: RNase Z family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P42980
- KEGG entry: [2]
- E.C. number: 4.2.3.3
Additional information
Expression and regulation
- Additional information:
Biological materials
- Expression vector:
- pGP1301 (N-terminal Strep-tag, purification from E. coli, in pGP172), available in Stülke lab
- pGP1180 (N-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP380), available in Stülke lab
- pGP1181 (C-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP382), available in Stülke lab
- pGP2207 (N-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP1459), available in Stülke lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Görke lab
- Antibody:
Labs working on this gene/protein
Boris Görke, University of Göttingen, Germany Homepage
Your additional remarks
References
Ahmed Gaballa, Haike Antelmann, Chris J Hamilton, John D Helmann
Regulation of Bacillus subtilis bacillithiol biosynthesis operons by Spx.
Microbiology (Reading): 2013, 159(Pt 10);2025-2035
[PubMed:23894131]
[WorldCat.org]
[DOI]
(I p)
Rachit Jain, Yajun Yan
Dehydratase mediated 1-propanol production in metabolically engineered Escherichia coli.
Microb Cell Fact: 2011, 10;97
[PubMed:22074179]
[WorldCat.org]
[DOI]
(I e)
Jens J Landmann, Ricarda A Busse, Jan-Hendrik Latz, Kalpana D Singh, Jörg Stülke, Boris Görke
Crh, the paralogue of the phosphocarrier protein HPr, controls the methylglyoxal bypass of glycolysis in Bacillus subtilis.
Mol Microbiol: 2011, 82(3);770-87
[PubMed:21992469]
[WorldCat.org]
[DOI]
(I p)
Ahmed Gaballa, Gerald L Newton, Haike Antelmann, Derek Parsonage, Heather Upton, Mamta Rawat, Al Claiborne, Robert C Fahey, John D Helmann
Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli.
Proc Natl Acad Sci U S A: 2010, 107(14);6482-6
[PubMed:20308541]
[WorldCat.org]
[DOI]
(I p)
D Saadat, D H Harrison
The crystal structure of methylglyoxal synthase from Escherichia coli.
Structure: 1999, 7(3);309-17
[PubMed:10368300]
[WorldCat.org]
[DOI]
(P p)