MetI
- Description: O-succinylhomoserine lyase (L-cysteine, H2S, methanethiol, elimination)
| Gene name | metI |
| Synonyms | yjcI |
| Essential | no |
| Product | O-succinylhomoserine lyase (L-cysteine, H2S, methanethiol, elimination) |
| Function | biosynthesis of methionine |
| Gene expression levels in SubtiExpress: metI | |
| Metabolic function and regulation of this protein in SubtiPathways: Cys, Met & Sulfate assimilation | |
| MW, pI | 41 kDa, 5.019 |
| Gene length, protein length | 1119 bp, 373 aa |
| Immediate neighbours | yjcH, metC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 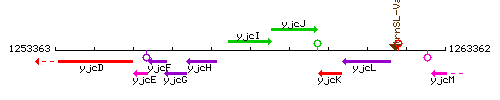
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed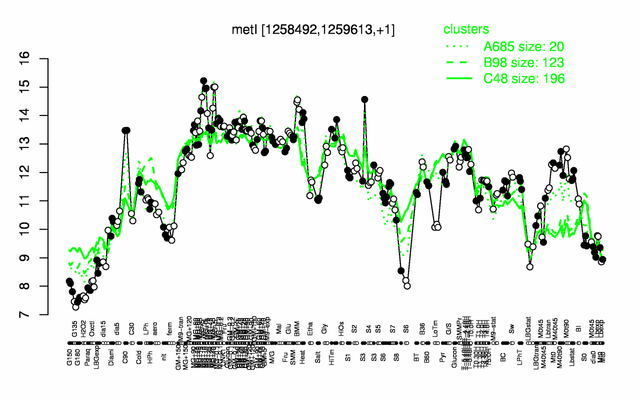
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU11870
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O31631
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism: S-box: transcription termination/ antitermination, the S-box riboswitch binds S-adenosylmethionine resulting in termination PubMed
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)
Jerneja Tomsic, Brooke A McDaniel, Frank J Grundy, Tina M Henkin
Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro.
J Bacteriol: 2008, 190(3);823-33
[PubMed:18039762]
[WorldCat.org]
[DOI]
(I p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
Sandrine Auger, W H Yuen, Antoine Danchin, Isabelle Martin-Verstraete
The metIC operon involved in methionine biosynthesis in Bacillus subtilis is controlled by transcription antitermination.
Microbiology (Reading): 2002, 148(Pt 2);507-518
[PubMed:11832514]
[WorldCat.org]
[DOI]
(P p)
F J Grundy, T M Henkin
The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria.
Mol Microbiol: 1998, 30(4);737-49
[PubMed:10094622]
[WorldCat.org]
[DOI]
(P p)