Difference between revisions of "MetA"
| Line 55: | Line 55: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU21910&redirect=T BSU21910] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/metA.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/metA.html] | ||
| Line 89: | Line 90: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU21910&redirect=T BSU21910] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2GHR 2GHR] (from ''Bacillus cereus'', 60% identity, 77% similarity) {{PubMed|17546672}} | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2GHR 2GHR] (from ''Bacillus cereus'', 60% identity, 77% similarity) {{PubMed|17546672}} | ||
Revision as of 14:00, 2 April 2014
- Description: homoserine O-succinyltransferase
| Gene name | metA |
| Synonyms | metB |
| Essential | no |
| Product | homoserine O-succinyltransferase |
| Function | biosynthesis of methionine |
| Gene expression levels in SubtiExpress: metA | |
| Metabolic function and regulation of this protein in SubtiPathways: metA | |
| MW, pI | 25 kDa, 6.414 |
| Gene length, protein length | 672 bp, 224 aa |
| Immediate neighbours | bsaA, ugtP |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 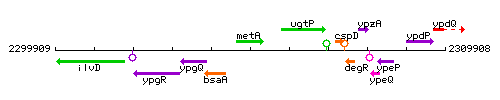
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed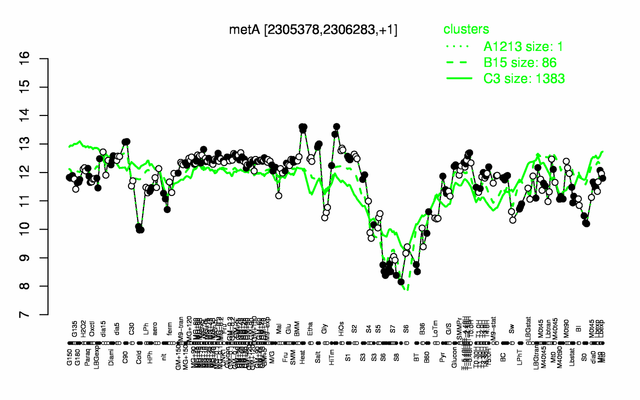
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids cell envelope stress proteins (controlled by SigM, V, W, X, Y)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU21910
Phenotypes of a mutant
Database entries
- BsubCyc: BSU21910
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Succinyl-CoA + L-homoserine = CoA + O-succinyl-L-homoserine (according to Swiss-Prot) O(4)-succinyl-L-homoserine + L-cysteine = L-cystathionine + succinate (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU21910
- UniProt: P54167
- KEGG entry: [3]
- E.C. number: 2.3.1.46 8 2.5.1.48]
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Chloe Zubieta, S Sri Krishna, Daniel McMullan, Mitchell D Miller, Polat Abdubek, Sanjay Agarwalla, Eileen Ambing, Tamara Astakhova, Herbert L Axelrod, Dennis Carlton, Hsiu-Ju Chiu, Thomas Clayton, Marc Deller, Michael DiDonato, Lian Duan, Marc-André Elsliger, Slawomir K Grzechnik, Joanna Hale, Eric Hampton, Gye Won Han, Justin Haugen, Lukasz Jaroszewski, Kevin K Jin, Heath E Klock, Mark W Knuth, Eric Koesema, Abhinav Kumar, David Marciano, Andrew T Morse, Edward Nigoghossian, Silvya Oommachen, Ron Reyes, Christopher L Rife, Henry van den Bedem, Dana Weekes, Aprilfawn White, Qingping Xu, Keith O Hodgson, John Wooley, Ashley M Deacon, Adam Godzik, Scott A Lesley, Ian A Wilson
Crystal structure of homoserine O-succinyltransferase from Bacillus cereus at 2.4 A resolution.
Proteins: 2007, 68(4);999-1005
[PubMed:17546672]
[WorldCat.org]
[DOI]
(I p)