Difference between revisions of "Mdh"
| Line 31: | Line 31: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=mdh_2978734_2979672_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:mdh_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=mdh_2978734_2979672_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:mdh_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU29120]] |
|- | |- | ||
|} | |} | ||
Revision as of 14:15, 16 May 2013
- Description: malate dehydrogenase
| Gene name | mdh |
| Synonyms | citH |
| Essential | no |
| Product | malate dehydrogenase |
| Function | TCA cycle |
| Gene expression levels in SubtiExpress: mdh | |
| Interactions involving this protein in SubtInteract: Mdh | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 33 kDa, 4.727 |
| Gene length, protein length | 936 bp, 312 aa |
| Immediate neighbours | phoP, icd |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 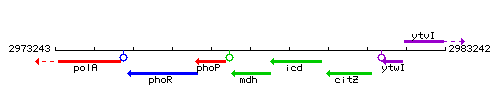
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed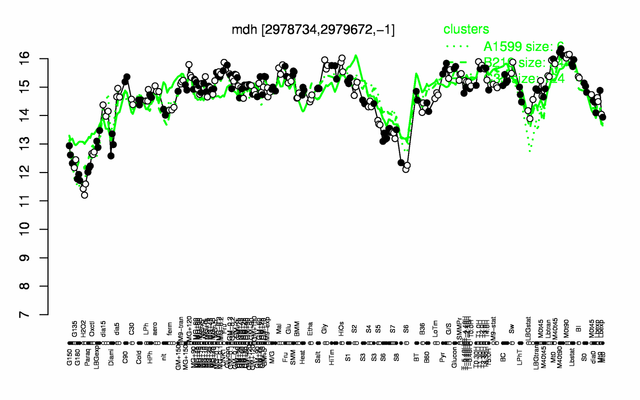
| |
Contents
Categories containing this gene/protein
carbon core metabolism, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29120
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (S)-malate + NAD+ = oxaloacetate + NADH (according to Swiss-Prot)
- Protein family: MDH type 3 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Reversible Michaelis-Menten PubMed
- Domains:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot), membrane associated PubMed
Database entries
- Structure: 1EMD (E.coli)
- UniProt: P49814
- KEGG entry: [3]
- E.C. number: 1.1.1.37
Additional information
- The enzyme is a tetramer PubMed
- extensive information on the structure and enzymatic properties of Mdh can be found at Proteopedia
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP719(spc) & GP1150(spc), available in Stülke lab
- Expression vector:
- pGP385: for expression, purification in E. coli with N-terminal His-tag, in pWH844, available in Jörg Stülke's lab
- pGP1123 (N-terminal Strep-tag, for SPINE, purification from B. subtilis, in pGP380) (available in Jörg Stülke's lab)
- pGP1755 (expression / purification of Mdh-S149A, with N-terminal His-tag from E. coli, in pWH844), available in Jörg Stülke's lab
- pGP1764 (for expression, purification in E. coli with N-terminal Strep-tag, in pGP172, available in Jörg Stülke's lab)
- GP1438(mdh-Strep (spc)) & GP1440(mdh-Strep (cat)), purification from B. subtilis, for SPINE, available in Stülke lab
- lacZ fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
- FLAG-tag construct:
- GP1130 (spc, based on pGP1331), available in Jörg Stülke's lab
Labs working on this gene/protein
Your additional remarks
References
Frederik M Meyer, Jörg Stülke
Malate metabolism in Bacillus subtilis: distinct roles for three classes of malate-oxidizing enzymes.
FEMS Microbiol Lett: 2013, 339(1);17-22
[PubMed:23136871]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Frederik M Meyer, Jan Gerwig, Elke Hammer, Christina Herzberg, Fabian M Commichau, Uwe Völker, Jörg Stülke
Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon.
Metab Eng: 2011, 13(1);18-27
[PubMed:20933603]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Hyun-Jin Kim, Agnes Roux, Abraham L Sonenshein
Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes.
Mol Microbiol: 2002, 45(1);179-90
[PubMed:12100558]
[WorldCat.org]
[DOI]
(P p)
C Jourlin-Castelli, N Mani, M M Nakano, A L Sonenshein
CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis.
J Mol Biol: 2000, 295(4);865-78
[PubMed:10656796]
[WorldCat.org]
[DOI]
(P p)
S Jin, M De Jesús-Berríos, A L Sonenshein
A Bacillus subtilis malate dehydrogenase gene.
J Bacteriol: 1996, 178(2);560-3
[PubMed:8550482]
[WorldCat.org]
[DOI]
(P p)
S Jin, A L Sonenshein
Transcriptional regulation of Bacillus subtilis citrate synthase genes.
J Bacteriol: 1994, 176(15);4680-90
[PubMed:8045899]
[WorldCat.org]
[DOI]
(P p)
A K Tyagi, F A Siddiqui, T A Venkitasubramanian
Studies on the purification and characterization of malate dehydrogenase from Mycobacterium phlei.
Biochim Biophys Acta: 1977, 485(2);255-67
[PubMed:922015]
[WorldCat.org]
[DOI]
(P p)
A YOSHIDA
ENZYMIC PROPERTIES OF MALATE DEHYDROGENASE OF BACILLUS SUBTILIS.
J Biol Chem: 1965, 240;1118-24
[PubMed:14284712]
[WorldCat.org]
(P p)