Difference between revisions of "Mdh"
| Line 32: | Line 32: | ||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| + | |||
| + | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[carbon core metabolism]]}}, | ||
| + | {{SubtiWiki category|[[membrane proteins]]}}, | ||
| + | {{SubtiWiki category|[[phosphoproteins]]}} | ||
| + | |||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[CcpA regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[CcpC regulon]]}} | ||
=The gene= | =The gene= | ||
| Line 51: | Line 60: | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
Revision as of 22:20, 8 December 2010
- Description: malate dehydrogenase
| Gene name | mdh |
| Synonyms | citH |
| Essential | no |
| Product | malate dehydrogenase |
| Function | TCA cycle |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 33 kDa, 4.727 |
| Gene length, protein length | 936 bp, 312 aa |
| Immediate neighbours | phoP, icd |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 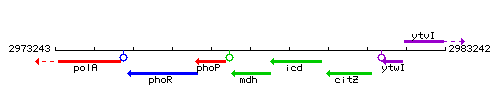
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
carbon core metabolism, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29120
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (S)-malate + NAD+ = oxaloacetate + NADH (according to Swiss-Prot)
- Protein family: MDH type 3 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Reversible Michaelis-Menten PubMed
- Domains:
- Modification: phosphorylation on Ser-149 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot), membrane associated PubMed
Database entries
- Structure: 1EMD (E.coli)
- UniProt: P49814
- KEGG entry: [3]
- E.C. number: 1.1.1.37
Additional information
The enzyme is a tetramer PubMed
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP719 (spc), available in Stülke lab
- Expression vector:
- pGP1123 (N-terminal Strep-tag, for SPINE, purification from B. subtilis, in pGP380) (available in Stülke lab)
- for expression, purification in E. coli with N-terminal His-tag, in pWH844: pGP385, available in Stülke lab
- pGP1755 (expression / purification of Mdh-S149A, with N-terminal His-tag from E. coli, in pWH844), available in Stülke lab
- pGP1764 (for expression, purification in E. coli with N-terminal Strep-tag, in pGP172, available in Stülke lab)
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Frederik M Meyer, Jan Gerwig, Elke Hammer, Christina Herzberg, Fabian M Commichau, Uwe Völker, Jörg Stülke
Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon.
Metab Eng: 2011, 13(1);18-27
[PubMed:20933603]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Hyun-Jin Kim, Agnes Roux, Abraham L Sonenshein
Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes.
Mol Microbiol: 2002, 45(1);179-90
[PubMed:12100558]
[WorldCat.org]
[DOI]
(P p)
C Jourlin-Castelli, N Mani, M M Nakano, A L Sonenshein
CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis.
J Mol Biol: 2000, 295(4);865-78
[PubMed:10656796]
[WorldCat.org]
[DOI]
(P p)
S Jin, M De Jesús-Berríos, A L Sonenshein
A Bacillus subtilis malate dehydrogenase gene.
J Bacteriol: 1996, 178(2);560-3
[PubMed:8550482]
[WorldCat.org]
[DOI]
(P p)
S Jin, A L Sonenshein
Transcriptional regulation of Bacillus subtilis citrate synthase genes.
J Bacteriol: 1994, 176(15);4680-90
[PubMed:8045899]
[WorldCat.org]
[DOI]
(P p)
A K Tyagi, F A Siddiqui, T A Venkitasubramanian
Studies on the purification and characterization of malate dehydrogenase from Mycobacterium phlei.
Biochim Biophys Acta: 1977, 485(2);255-67
[PubMed:922015]
[WorldCat.org]
[DOI]
(P p)
A YOSHIDA
ENZYMIC PROPERTIES OF MALATE DEHYDROGENASE OF BACILLUS SUBTILIS.
J Biol Chem: 1965, 240;1118-24
[PubMed:14284712]
[WorldCat.org]
(P p)