Difference between revisions of "McsA"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Nobse) |
|||
| Line 51: | Line 51: | ||
* '''Locus tag:''' BSU00840 | * '''Locus tag:''' BSU00840 | ||
| − | |||
| − | |||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
Revision as of 17:30, 27 January 2012
| Gene name | mcsA |
| Synonyms | yacH |
| Essential | no |
| Product | activator of McsB kinase activity |
| Function | control of CtsR activity |
| Interactions involving this protein in SubtInteract: McsA | |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 20 kDa, 6.624 |
| Gene length, protein length | 555 bp, 185 aa |
| Immediate neighbours | ctsR, mcsB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 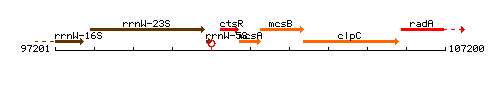
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription factors and their control, sporulation proteins, general stress proteins (controlled by SigB), heat shock proteins
This gene is a member of the following regulons
CtsR regulon, SigB regulon, SigF regulon
The gene
Basic information
- Locus tag: BSU00840
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P37569
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant: mcsA::aphA3 available from the Gerth lab
- Expression vector: for expression, purification in E. coli with N-terminal His-tag, pRSETA available in Gerth lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in Gerth lab
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Jeanette Hahn, Naomi Kramer, Kenneth Briley, David Dubnau
McsA and B mediate the delocalization of competence proteins from the cell poles of Bacillus subtilis.
Mol Microbiol: 2009, 72(1);202-15
[PubMed:19226326]
[WorldCat.org]
[DOI]
(I p)
Stephanie T Wang, Barbara Setlow, Erin M Conlon, Jessica L Lyon, Daisuke Imamura, Tsutomu Sato, Peter Setlow, Richard Losick, Patrick Eichenberger
The forespore line of gene expression in Bacillus subtilis.
J Mol Biol: 2006, 358(1);16-37
[PubMed:16497325]
[WorldCat.org]
[DOI]
(P p)
Pekka Varmanen, Finn K Vogensen, Karin Hammer, Airi Palva, Hanne Ingmer
ClpE from Lactococcus lactis promotes repression of CtsR-dependent gene expression.
J Bacteriol: 2003, 185(17);5117-24
[PubMed:12923084]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
E Krüger, D Zühlke, E Witt, H Ludwig, M Hecker
Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor.
EMBO J: 2001, 20(4);852-63
[PubMed:11179229]
[WorldCat.org]
[DOI]
(P p)
I Derré, G Rapoport, T Msadek
CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria.
Mol Microbiol: 1999, 31(1);117-31
[PubMed:9987115]
[WorldCat.org]
[DOI]
(P p)
E Krüger, T Msadek, M Hecker
Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon.
Mol Microbiol: 1996, 20(4);713-23
[PubMed:8793870]
[WorldCat.org]
[DOI]
(P p)