Difference between revisions of "Map"
| Line 60: | Line 60: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU01380&redirect=T BSU01380] | ||
* '''DBTBS entry:''' no entry | * '''DBTBS entry:''' no entry | ||
| Line 101: | Line 102: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU01380&redirect=T BSU01380] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2MAT 2MAT] (from ''Escherichia coli'', 46% identity, 65% similarity) {{PubMed|10387007}} | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2MAT 2MAT] (from ''Escherichia coli'', 46% identity, 65% similarity) {{PubMed|10387007}} | ||
Revision as of 12:50, 2 April 2014
- Description: methionine aminopeptidase
| Gene name | map |
| Synonyms | mapA |
| Essential | yes PubMed |
| Product | methionine aminopeptidase |
| Function | removal of N-terminal methionine from nascent proteins |
| Gene expression levels in SubtiExpress: map | |
| Interactions involving this protein in SubtInteract: Map | |
| Metabolic function and regulation of this protein in SubtiPathways: map | |
| MW, pI | 27 kDa, 6.393 |
| Gene length, protein length | 744 bp, 248 aa |
| Immediate neighbours | adk, ybzG |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 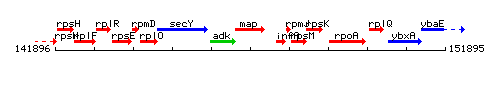
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed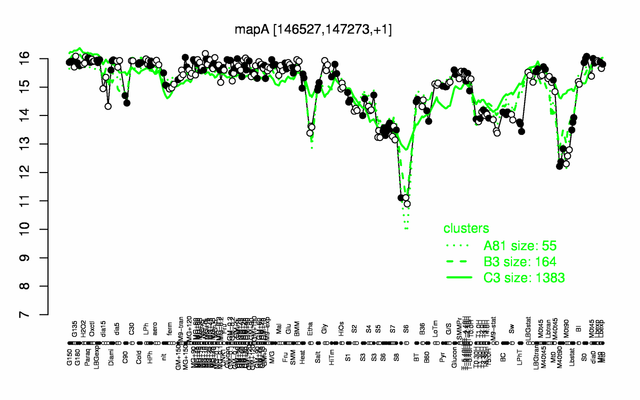
| |
Contents
Categories containing this gene/protein
translation, protein modification, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01380
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU01380
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Release of N-terminal amino acids, preferentially methionine, from peptides and arylamides (according to Swiss-Prot)
- Protein family: peptidase M24A family (according to Swiss-Prot)
- Paralogous protein(s): YflG
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU01380
- UniProt: P19994
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information: strongly expressed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Arzu Sandikci, Felix Gloge, Michael Martinez, Matthias P Mayer, Rebecca Wade, Bernd Bukau, Günter Kramer
Dynamic enzyme docking to the ribosome coordinates N-terminal processing with polypeptide folding.
Nat Struct Mol Biol: 2013, 20(7);843-50
[PubMed:23770820]
[WorldCat.org]
[DOI]
(I p)
CongHui You, HongYan Lu, Agnieszka Sekowska, Gang Fang, YiPing Wang, Anne-Marie Gilles, Antoine Danchin
The two authentic methionine aminopeptidase genes are differentially expressed in Bacillus subtilis.
BMC Microbiol: 2005, 5;57
[PubMed:16207374]
[WorldCat.org]
[DOI]
(I e)
J W Suh, S A Boylan, S H Oh, C W Price
Genetic and transcriptional organization of the Bacillus subtilis spc-alpha region.
Gene: 1996, 169(1);17-23
[PubMed:8635744]
[WorldCat.org]
[DOI]
(P p)