Difference between revisions of "LytE"
| Line 39: | Line 39: | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| − | {{SubtiWiki category|[[cell wall degradation/ turnover]]}} | + | {{SubtiWiki category|[[cell wall degradation/ turnover]]}}, |
| + | {{SubtiWiki category|[[cell wall synthesis]]}} | ||
| + | |||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 57: | Line 59: | ||
* growth defect at high temperature {{PubMed|21541672}} | * growth defect at high temperature {{PubMed|21541672}} | ||
* inactivation of ''[[lytE]]'' strongly restores beta-lactam resistance in a ''[[sigM]]'' mutant by delaying cell lysis {{PubMed|22211522}} | * inactivation of ''[[lytE]]'' strongly restores beta-lactam resistance in a ''[[sigM]]'' mutant by delaying cell lysis {{PubMed|22211522}} | ||
| + | * a ''[[lytE]]'' mutation is synthetically lethal with ''[[ftsE]]'' and ''[[ftsX]]'' mutation (due to a lack of autolysin activity) {{PubMed|xxx}} | ||
| + | * a ''[[lytE]]'' mutation increases the cell separation defect of a ''[[lytF]]'' mutant {{PubMed|xxx}} | ||
=== Database entries === | === Database entries === | ||
| Line 118: | Line 122: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=lytE_1018998_1020002_1 lytE] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=lytE_1018998_1020002_1 lytE] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|9573210}}, [[SigH]] {{PubMed|9573210}}, [[SigI]] {{PubMed|21541672}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|9573210}}, [[SigH]] {{PubMed|9573210}}, [[SigI]] {{PubMed|21541672}} |
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 12:33, 16 July 2013
- Description: cell wall hydrolase (major autolysin), D,L-endopeptidase-type autolysin
| Gene name | lytE |
| Synonyms | papQ, cwlF |
| Essential | no |
| Product | cell wall hydrolase (major autolysin),endopeptidase-type autolysin |
| Function | major autolysin, cell separation, cell proliferation |
| Gene expression levels in SubtiExpress: lytE | |
| MW, pI | 37 kDa, 10.713 |
| Gene length, protein length | 1029 bp, 343 aa |
| Immediate neighbours | phoA, citR |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 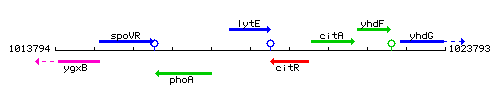
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed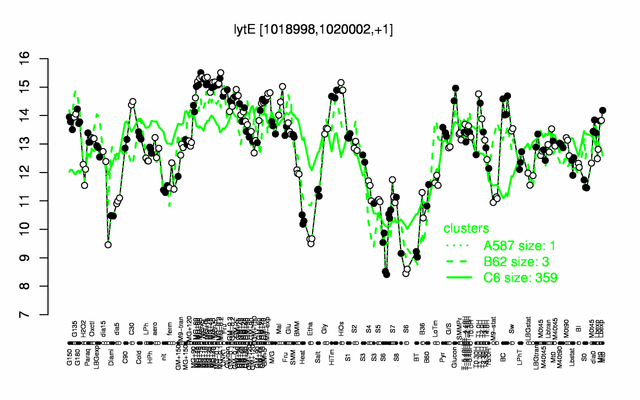
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover, cell wall synthesis
This gene is a member of the following regulons
SigH regulon, SigI regulon, Spo0A regulon, WalR regulon
The gene
Basic information
- Locus tag: BSU09420
Phenotypes of a mutant
- a cwlO lytE mutant is not viable PubMed
- growth defect at high temperature PubMed
- inactivation of lytE strongly restores beta-lactam resistance in a sigM mutant by delaying cell lysis PubMed
- a lytE mutation is synthetically lethal with ftsE and ftsX mutation (due to a lack of autolysin activity) PubMed
- a lytE mutation increases the cell separation defect of a lytF mutant PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: nlpC/p60 family (according to Swiss-Prot)
- Paralogous protein(s): the C-terminal D,L-endopeptidase domains of LytE, LytF, CwlS, and CwlO exhibit strong sequence similarity
Extended information on the protein
- Kinetic information:
- Domains:
- C-terminal D,L-endopeptidase domain PubMed
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P54421
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: lytE PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Additional reviews: PubMed
Original publications
Additional publications: PubMed
Masayuki Hashimoto, Seika Ooiwa, Junichi Sekiguchi
Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall.
J Bacteriol: 2012, 194(4);796-803
[PubMed:22139507]
[WorldCat.org]
[DOI]
(I p)
Chi-Ling Tseng, Jung-Tze Chen, Ju-Hui Lin, Wan-Zhen Huang, Gwo-Chyuan Shaw
Genetic evidence for involvement of the alternative sigma factor SigI in controlling expression of the cell wall hydrolase gene lytE and contribution of LytE to heat survival of Bacillus subtilis.
Arch Microbiol: 2011, 193(9);677-85
[PubMed:21541672]
[WorldCat.org]
[DOI]
(I p)
Paola Bisicchia, David Noone, Efthimia Lioliou, Alistair Howell, Sarah Quigley, Thomas Jensen, Hanne Jarmer, Kevin M Devine
The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis.
Mol Microbiol: 2007, 65(1);180-200
[PubMed:17581128]
[WorldCat.org]
[DOI]
(P p)
Rut Carballido-López, Alex Formstone, Ying Li, S Dusko Ehrlich, Philippe Noirot, Jeff Errington
Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE.
Dev Cell: 2006, 11(3);399-409
[PubMed:16950129]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Hiroki Yamamoto, Shin-ichirou Kurosawa, Junichi Sekiguchi
Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases.
J Bacteriol: 2003, 185(22);6666-77
[PubMed:14594841]
[WorldCat.org]
[DOI]
(P p)
R Ohnishi, S Ishikawa, J Sekiguchi
Peptidoglycan hydrolase LytF plays a role in cell separation with CwlF during vegetative growth of Bacillus subtilis.
J Bacteriol: 1999, 181(10);3178-84
[PubMed:10322020]
[WorldCat.org]
[DOI]
(P p)
S Ishikawa, Y Hara, R Ohnishi, J Sekiguchi
Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis.
J Bacteriol: 1998, 180(9);2549-55
[PubMed:9573210]
[WorldCat.org]
[DOI]
(P p)
P Margot, M Wahlen, A Gholamhoseinian, P Piggot, D Karamata
The lytE gene of Bacillus subtilis 168 encodes a cell wall hydrolase.
J Bacteriol: 1998, 180(3);749-52
[PubMed:9457885]
[WorldCat.org]
[DOI]
(P p)