LysA
- Description: diaminopimelate decarboxylase
| Gene name | lysA |
| Synonyms | |
| Essential | no |
| Product | diaminopimelate decarboxylase |
| Function | biosynthesis of lysine |
| Gene expression levels in SubtiExpress: lysA | |
| Metabolic function and regulation of this protein in SubtiPathways: lysA | |
| MW, pI | 48 kDa, 5.007 |
| Gene length, protein length | 1317 bp, 439 aa |
| Immediate neighbours | ypuA, spoVAF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 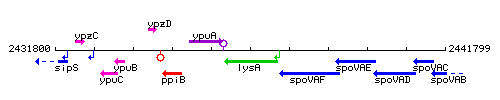
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed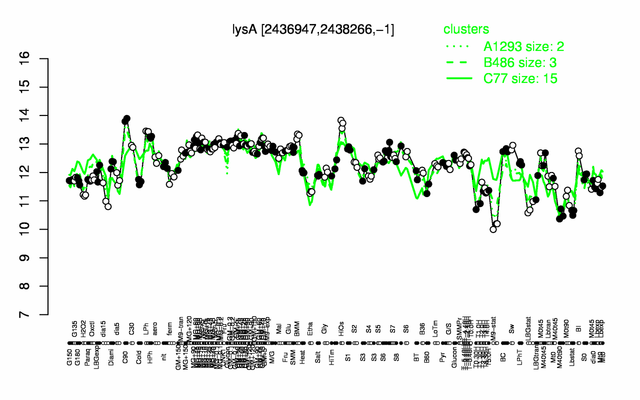
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU23380
Phenotypes of a mutant
- auxotrophic for lysine PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Meso-2,6-diaminoheptanedioate = L-lysine + CO2 (according to Swiss-Prot)
- Protein family: Orn/Lys/Arg decarboxylase class-II family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- UniProt: P23630
- KEGG entry: [3]
- E.C. number: 4.1.1.20
Additional information
Expression and regulation
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Leif Steil, Mónica Serrano, Adriano O Henriques, Uwe Völker
Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 2);399-420
[PubMed:15699190]
[WorldCat.org]
[DOI]
(P p)
Alain Brans, Patrice Filée, Andy Chevigné, Aurore Claessens, Bernard Joris
New integrative method to generate Bacillus subtilis recombinant strains free of selection markers.
Appl Environ Microbiol: 2004, 70(12);7241-50
[PubMed:15574923]
[WorldCat.org]
[DOI]
(P p)
I Bagyan, J Hobot, S Cutting
A compartmentalized regulator of developmental gene expression in Bacillus subtilis.
J Bacteriol: 1996, 178(15);4500-7
[PubMed:8755877]
[WorldCat.org]
[DOI]
(P p)
V Azevedo, A Sorokin, S D Ehrlich, P Serror
The transcriptional organization of the Bacillus subtilis 168 chromosome region between the spoVAF and serA genetic loci.
Mol Microbiol: 1993, 10(2);397-405
[PubMed:7934830]
[WorldCat.org]
[DOI]
(P p)
B Moldover, P J Piggot, M D Yudkin
Identification of the promoter and the transcriptional start site of the spoVA operon of Bacillus subtilis and Bacillus licheniformis.
J Gen Microbiol: 1991, 137(3);527-31
[PubMed:1903432]
[WorldCat.org]
[DOI]
(P p)