LutB
- Description: lactate catabolic enzyme
| Gene name | lutB |
| Synonyms | yvfW |
| Essential | no |
| Product | lactate oxidase |
| Function | lactate utilization |
| Gene expression levels in SubtiExpress: lutB | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 53 kDa, 5.848 |
| Gene length, protein length | 1437 bp, 479 aa |
| Immediate neighbours | lutC, lutA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 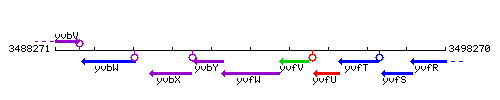
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed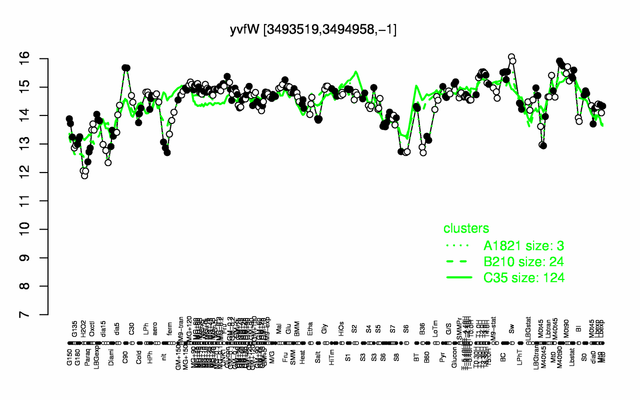
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, phosphoproteins
This gene is a member of the following regulons
FbpB regulon, FsrA regulon, LutR regulon, SinR regulon
The gene
Basic information
- Locus tag: BSU34040
Phenotypes of a mutant
no growth with lactate as the single carbon source PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: oxidation of lactate to pyruvate PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-33 PubMed
- Cofactor(s): contains an iron-sulfur cluster
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O07021
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Richard Losick, Harvard Univ., Cambridge, USA homepage
Your additional remarks
References
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Gregory T Smaldone, Haike Antelmann, Ahmed Gaballa, John D Helmann
The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases.
J Bacteriol: 2012, 194(10);2586-93
[PubMed:22427629]
[WorldCat.org]
[DOI]
(I p)
Gregory T Smaldone, Olga Revelles, Ahmed Gaballa, Uwe Sauer, Haike Antelmann, John D Helmann
A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism.
J Bacteriol: 2012, 194(10);2594-605
[PubMed:22389480]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Roberto Kolter, Richard Losick
A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation.
J Bacteriol: 2009, 191(8);2423-30
[PubMed:19201793]
[WorldCat.org]
[DOI]
(I p)
Ahmed Gaballa, Haike Antelmann, Claudio Aguilar, Sukhjit K Khakh, Kyung-Bok Song, Gregory T Smaldone, John D Helmann
The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins.
Proc Natl Acad Sci U S A: 2008, 105(33);11927-32
[PubMed:18697947]
[WorldCat.org]
[DOI]
(I p)
Frances Chu, Daniel B Kearns, Steven S Branda, Roberto Kolter, Richard Losick
Targets of the master regulator of biofilm formation in Bacillus subtilis.
Mol Microbiol: 2006, 59(4);1216-28
[PubMed:16430695]
[WorldCat.org]
[DOI]
(P p)