Difference between revisions of "LpdV"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || utilization of branched-chain keto acids | |style="background:#ABCDEF;" align="center"|'''Function''' || utilization of branched-chain keto acids | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU24060 lpdV] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_synthesis.html Lipid synthesis], [http://subtiwiki.uni-goettingen.de/pathways/ile_val_leu.html Ile, Leu, Val]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_synthesis.html Lipid synthesis], [http://subtiwiki.uni-goettingen.de/pathways/ile_val_leu.html Ile, Leu, Val]''' | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[bkdAA]]'', ''[[buk]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[bkdAA]]'', ''[[buk]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU24060 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU24060 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU24060 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:lpdV_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:lpdV_context.gif]] | ||
Revision as of 13:14, 13 May 2013
- Description: 2-oxoisovalerate dehydrogenase (E3 subunit, dihydrolipoamide dehydrogenase)
| Gene name | lpdV |
| Synonyms | yqiV, bkd |
| Essential | no |
| Product | 2-oxoisovalerate dehydrogenase (E3 subunit, dihydrolipoamide dehydrogenase) |
| Function | utilization of branched-chain keto acids |
| Gene expression levels in SubtiExpress: lpdV | |
| Metabolic function and regulation of this protein in SubtiPathways: Lipid synthesis, Ile, Leu, Val | |
| MW, pI | 48 kDa, 4.893 |
| Gene length, protein length | 1371 bp, 457 aa |
| Immediate neighbours | bkdAA, buk |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 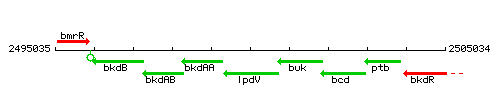
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
BkdR regulon, CodY regulon, SigL regulon
The gene
Basic information
- Locus tag: BSU24060
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein N(6)-(dihydrolipoyl)lysine + NAD+ = protein N(6)-(lipoyl)lysine + NADH (according to Swiss-Prot)
- Protein family: class-I pyridine nucleotide-disulfide oxidoreductase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 2YQU (from Thermus thermophilus (hb8 mutant), 42% identity, 55% similarity)
- UniProt: P54533
- KEGG entry: [3]
- E.C. number: 1.8.1.4
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
M Nickel, G Homuth, C Böhnisch, U Mäder, T Schweder
Cold induction of the Bacillus subtilis bkd operon is mediated by increased mRNA stability.
Mol Genet Genomics: 2004, 272(1);98-107
[PubMed:15241682]
[WorldCat.org]
[DOI]
(P p)
Ken-ichi Yoshida, Hirotake Yamaguchi, Masaki Kinehara, Yo-hei Ohki, Yoshiko Nakaura, Yasutaro Fujita
Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box.
Mol Microbiol: 2003, 49(1);157-65
[PubMed:12823818]
[WorldCat.org]
[DOI]
(P p)
Tanja Kaan, Georg Homuth, Ulrike Mäder, Julia Bandow, Thomas Schweder
Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response.
Microbiology (Reading): 2002, 148(Pt 11);3441-3455
[PubMed:12427936]
[WorldCat.org]
[DOI]
(P p)
M Debarbouille, R Gardan, M Arnaud, G Rapoport
Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis.
J Bacteriol: 1999, 181(7);2059-66
[PubMed:10094682]
[WorldCat.org]
[DOI]
(P p)