Difference between revisions of "LipC"
| Line 29: | Line 29: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=lipC_462431_463072_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:lipC_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=lipC_462431_463072_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:lipC_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU04110]] |
|- | |- | ||
|} | |} | ||
Revision as of 12:34, 16 May 2013
- Description: phospholipase implicated in spore germination
| Gene name | lipC |
| Synonyms | ycsK |
| Essential | no |
| Product | spore coat phospholipase B |
| Function | spore germination |
| Gene expression levels in SubtiExpress: lipC | |
| Function and regulation of this protein in SubtiPathways: Phosphorelay | |
| MW, pI | 23 kDa, 5.839 |
| Gene length, protein length | 639 bp, 213 aa |
| Immediate neighbours | kipR, yczI |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 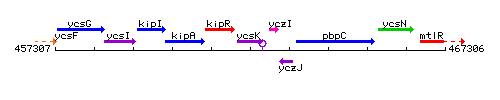
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed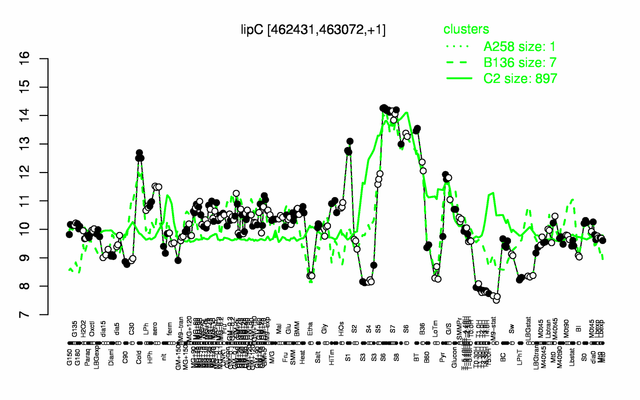
| |
Contents
Categories containing this gene/protein
utilization of lipids, sporulation proteins, germination
This gene is a member of the following regulons
KipR regulon, SigK regulon, TnrA regulon
The gene
Basic information
- Locus tag: BSU04110
Phenotypes of a mutant
defective in L-alanine-stimulated germination PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: cleaves the fatty acids at the sn-1 and sn-2 positions of phospholipids PubMed
- Protein family: 'GDSL' lipolytic enzyme family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- spore coat PubMed
Database entries
- Structure:
- UniProt: P42969
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Atsushi Masayama, Shiro Kato, Takuya Terashima, Anne Mølgaard, Hisashi Hemmi, Tohru Yoshimura, Ryuichi Moriyama
Bacillus subtilis spore coat protein LipC is a phospholipase B.
Biosci Biotechnol Biochem: 2010, 74(1);24-30
[PubMed:20057119]
[WorldCat.org]
[DOI]
(I p)
Atsushi Masayama, Ritsuko Kuwana, Hiromu Takamatsu, Hisashi Hemmi, Tohru Yoshimura, Kazuhito Watabe, Ryuichi Moriyama
A novel lipolytic enzyme, YcsK (LipC), located in the spore coat of Bacillus subtilis, is involved in spore germination.
J Bacteriol: 2007, 189(6);2369-75
[PubMed:17220230]
[WorldCat.org]
[DOI]
(P p)
L Wang, R Grau, M Perego, J A Hoch
A novel histidine kinase inhibitor regulating development in Bacillus subtilis.
Genes Dev: 1997, 11(19);2569-79
[PubMed:9334321]
[WorldCat.org]
[DOI]
(P p)