Difference between revisions of "LexA"
(→This gene is a member of the following regulons) |
(→Expression and regulation) |
||
| Line 100: | Line 100: | ||
* '''Operon:''' ''[[lexA]]'' {{PubMed|1657879}} | * '''Operon:''' ''[[lexA]]'' {{PubMed|1657879}} | ||
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 18:58, 20 May 2011
- Description: transcriptional repressor of the SOS regulon
| Gene name | lexA |
| Synonyms | dinR |
| Essential | no |
| Product | transcriptional repressor of the SOS regulon |
| Function | regulation of DNA damage repair |
| Function and regulation of this protein in SubtiPathways: Phosphorelay, Protein secretion | |
| MW, pI | 22 kDa, 5.155 |
| Gene length, protein length | 615 bp, 205 aa |
| Immediate neighbours | fosB, yneA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 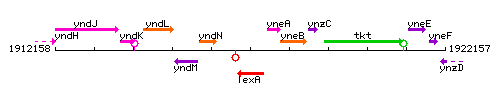
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, transcription factors and their control, general stress proteins (controlled by SigB)
This gene is a member of the following regulons
The LexA regulon
The gene
Basic information
- Locus tag: BSU17850
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Represses uvrB, dinB, dinC, recA genes and itself by binding to the 14 bp palindromic sequence 5'-CGAACNNNNGTTCG-3'. In the presence of single-stranded DNA, RecA interacts with LexA causing an autocatalytic cleavage which disrupts the DNA-binding part of LexA, leading to derepression of the SOS regulon and eventually DNA repair. (according to Swiss-Prot)
- Protein family: peptidase S24 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- UniProt: P31080
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Yoshikazu Kawai, Naotake Ogasawara
Bacillus subtilis EzrA and FtsL synergistically regulate FtsZ ring dynamics during cell division.
Microbiology (Reading): 2006, 152(Pt 4);1129-1141
[PubMed:16549676]
[WorldCat.org]
[DOI]
(P p)
Nora Au, Elke Kuester-Schoeck, Veena Mandava, Laura E Bothwell, Susan P Canny, Karen Chachu, Sierra A Colavito, Shakierah N Fuller, Eli S Groban, Laura A Hensley, Theresa C O'Brien, Amish Shah, Jessica T Tierney, Louise L Tomm, Thomas M O'Gara, Alexi I Goranov, Alan D Grossman, Charles M Lovett
Genetic composition of the Bacillus subtilis SOS system.
J Bacteriol: 2005, 187(22);7655-66
[PubMed:16267290]
[WorldCat.org]
[DOI]
(P p)
Y Luo, R A Pfuetzner, S Mosimann, M Paetzel, E A Frey, M Cherney, B Kim, J W Little, N C Strynadka
Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell: 2001, 106(5);585-94
[PubMed:11551506]
[WorldCat.org]
[DOI]
(P p)
C W Price, P Fawcett, H Cérémonie, N Su, C K Murphy, P Youngman
Genome-wide analysis of the general stress response in Bacillus subtilis.
Mol Microbiol: 2001, 41(4);757-74
[PubMed:11532142]
[WorldCat.org]
[DOI]
(P p)
K W Winterling, D Chafin, J J Hayes, J Sun, A S Levine, R E Yasbin, R Woodgate
The Bacillus subtilis DinR binding site: redefinition of the consensus sequence.
J Bacteriol: 1998, 180(8);2201-11
[PubMed:9555905]
[WorldCat.org]
[DOI]
(P p)
K W Winterling, A S Levine, R E Yasbin, R Woodgate
Characterization of DinR, the Bacillus subtilis SOS repressor.
J Bacteriol: 1997, 179(5);1698-703
[PubMed:9045831]
[WorldCat.org]
[DOI]
(P p)
M C Miller, J B Resnick, B T Smith, C M Lovett
The bacillus subtilis dinR gene codes for the analogue of Escherichia coli LexA. Purification and characterization of the DinR protein.
J Biol Chem: 1996, 271(52);33502-8
[PubMed:8969214]
[WorldCat.org]
(P p)
B J Haijema, D van Sinderen, K Winterling, J Kooistra, G Venema, L W Hamoen
Regulated expression of the dinR and recA genes during competence development and SOS induction in Bacillus subtilis.
Mol Microbiol: 1996, 22(1);75-85
[PubMed:8899710]
[WorldCat.org]
[DOI]
(P p)
A Raymond-Denise, N Guillen
Identification of dinR, a DNA damage-inducible regulator gene of Bacillus subtilis.
J Bacteriol: 1991, 173(22);7084-91
[PubMed:1657879]
[WorldCat.org]
[DOI]
(P p)