Difference between revisions of "HxlR"
(→Extended information on the protein) |
|||
| Line 80: | Line 80: | ||
* '''Effectors of protein activity:''' formaldehyde stimulates transcription activation by HxlR | * '''Effectors of protein activity:''' formaldehyde stimulates transcription activation by HxlR | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
Revision as of 14:46, 15 August 2011
| Gene name | hxlR |
| Synonyms | yckH |
| Essential | no |
| Product | transcription activator |
| Function | regulation of the ribulose monophosphate pathway |
| MW, pI | 14 kDa, 6.358 |
| Gene length, protein length | 360 bp, 120 aa |
| Immediate neighbours | hxlA, srfAA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 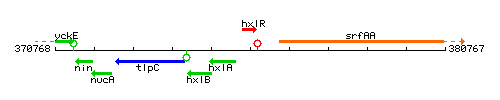
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription factors and their control, resistance against oxidative and electrophile stress
This gene is a member of the following regulons
The HxlR regulon:
The gene
Basic information
- Locus tag: BSU03470
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: formaldehyde stimulates transcription activation by HxlR
Database entries
- Structure:
- UniProt: P42406
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: hxlR (according to DBTBS)
- Regulation:
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
H Yasueda, Y Kawahara, S Sugimoto
Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression.
J Bacteriol: 1999, 181(23);7154-60
[PubMed:10572115]
[WorldCat.org]
[DOI]
(P p)