Difference between revisions of "HutU"
| Line 122: | Line 122: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 314 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 10:08, 17 April 2014
- Description: urocanase
| Gene name | hutU |
| Synonyms | |
| Essential | no |
| Product | urocanase |
| Function | histidine utilization |
| Gene expression levels in SubtiExpress: hutU | |
| Metabolic function and regulation of this protein in SubtiPathways: hutU | |
| MW, pI | 60 kDa, 5.664 |
| Gene length, protein length | 1656 bp, 552 aa |
| Immediate neighbours | hutH, hutI |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 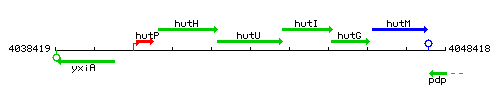
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed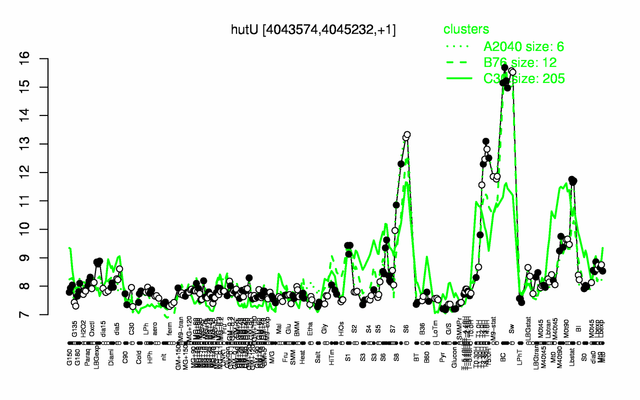
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
CcpA regulon, CodY regulon, HutP regulon
The gene
Basic information
- Locus tag: BSU39360
Phenotypes of a mutant
Database entries
- BsubCyc: BSU39360
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 3-(5-oxo-4,5-dihydro-3H-imidazol-4-yl)propanoate = urocanate + H2O (according to Swiss-Prot)
- Protein family: urocanase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU39360
- Structure: 2FKN
- UniProt: P25503
- KEGG entry: [3]
- E.C. number: 4.2.1.49
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (complex medium with amino acids, without glucose): 314 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Robert A Bender
Regulation of the histidine utilization (hut) system in bacteria.
Microbiol Mol Biol Rev: 2012, 76(3);565-84
[PubMed:22933560]
[WorldCat.org]
[DOI]
(I p)
Original publications
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Ken-Ichi Yoshida, Izumi Ishio, Eishi Nagakawa, Yoshiyuki Yamamoto, Mami Yamamoto, Yasutaro Fujita
Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome.
Microbiology (Reading): 2000, 146 ( Pt 3);573-579
[PubMed:10746760]
[WorldCat.org]
[DOI]
(P p)
J M Zalieckas, L V Wray, S H Fisher
trans-acting factors affecting carbon catabolite repression of the hut operon in Bacillus subtilis.
J Bacteriol: 1999, 181(9);2883-8
[PubMed:10217782]
[WorldCat.org]
[DOI]
(P p)
S H Fisher, K Rohrer, A E Ferson
Role of CodY in regulation of the Bacillus subtilis hut operon.
J Bacteriol: 1996, 178(13);3779-84
[PubMed:8682780]
[WorldCat.org]
[DOI]
(P p)
L V Wray, S H Fisher
Analysis of Bacillus subtilis hut operon expression indicates that histidine-dependent induction is mediated primarily by transcriptional antitermination and that amino acid repression is mediated by two mechanisms: regulation of transcription initiation and inhibition of histidine transport.
J Bacteriol: 1994, 176(17);5466-73
[PubMed:8071225]
[WorldCat.org]
[DOI]
(P p)