Difference between revisions of "Hom"
(→References) |
|||
| Line 146: | Line 146: | ||
=References= | =References= | ||
| − | <pubmed>12107147 18763711 3139660, 19258532 24163341 25777134 15378759</pubmed> | + | <pubmed>12107147 18763711 3139660, 19258532 24163341 25777134 15378759 25755103</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:35, 30 July 2015
- Description: homoserine dehydrogenase (NADPH)
| Gene name | hom |
| Synonyms | |
| Essential | no |
| Product | homoserine dehydrogenase (NADPH) |
| Function | biosynthesis of methionine and threonine |
| Gene expression levels in SubtiExpress: hom | |
| Metabolic function and regulation of this protein in SubtiPathways: hom | |
| MW, pI | 47 kDa, 4.9 |
| Gene length, protein length | 1299 bp, 433 aa |
| Immediate neighbours | thrC, yutH |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 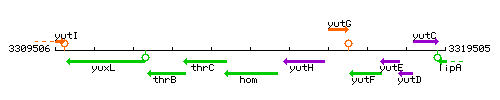
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed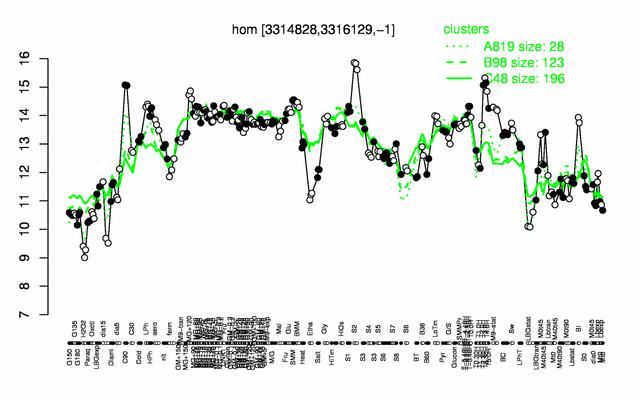
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, membrane proteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU32260
Phenotypes of a mutant
Database entries
- BsubCyc: BSU32260
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-homoserine + NAD(P)+ = L-aspartate 4-semialdehyde + NAD(P)H (according to Swiss-Prot)
- Protein family: homoserine dehydrogenase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity: subject to feedback inhibition PubMed
- Localization:
- membrane associated PubMed
Database entries
- BsubCyc: BSU32260
- Structure: 2EJW (from Thermus thermophilus hb8, 37% identity, 57% similarity)
- UniProt: P19582
- KEGG entry: [2]
- E.C. number: 1.1.1.3
Additional information
Expression and regulation
- Regulation:
- Additional information:
- subject to feedback inhibition PubMed
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 2167 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 1723 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 3436 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 2047 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 3239 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Fabian M Commichau, Ariane Alzinger, Rafael Sande, Werner Bretzel, Daniel R Reuß, Miriam Dormeyer, Bastien Chevreux, Jörg Schuldes, Rolf Daniel, Michiel Akeroyd, Markus Wyss, Hans-Peter Hohmann, Zoltán Prágai
Engineering Bacillus subtilis for the conversion of the antimetabolite 4-hydroxy-l-threonine to pyridoxine.
Metab Eng: 2015, 29;196-207
[PubMed:25777134]
[WorldCat.org]
[DOI]
(I p)
Nicolas Mirouze, Elena Bidnenko, Philippe Noirot, Sandrine Auger
Genome-wide mapping of TnrA-binding sites provides new insights into the TnrA regulon in Bacillus subtilis.
Microbiologyopen: 2015, 4(3);423-35
[PubMed:25755103]
[WorldCat.org]
[DOI]
(I p)
Allison Kriel, Shaun R Brinsmade, Jessica L Tse, Ashley K Tehranchi, Alycia N Bittner, Abraham L Sonenshein, Jue D Wang
GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes.
J Bacteriol: 2014, 196(1);189-201
[PubMed:24163341]
[WorldCat.org]
[DOI]
(I p)
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
C Parsot, G N Cohen
Cloning and nucleotide sequence of the Bacillus subtilis hom gene coding for homoserine dehydrogenase. Structural and evolutionary relationships with Escherichia coli aspartokinases-homoserine dehydrogenases I and II.
J Biol Chem: 1988, 263(29);14654-60
[PubMed:3139660]
[WorldCat.org]
(P p)