Difference between revisions of "HisC"
| (31 intermediate revisions by 6 users not shown) | |||
| Line 4: | Line 4: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''hisC '' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' aroJ'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || biosynthesis of aromatic amino acids | |style="background:#ABCDEF;" align="center"|'''Function''' || biosynthesis of aromatic amino acids | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU22620 hisC] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=hisC hisC]''' | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 39 kDa, 5.005 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 39 kDa, 5.005 | ||
| Line 20: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[tyrA]]'', ''[[trpA]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[tyrA]]'', ''[[trpA]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU22620 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU22620 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU22620 DNA_with_flanks] |
|- | |- | ||
|- | |- | ||
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:hisC_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:hisC_context.gif]] | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=hisC_2370415_2371497_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:hisC_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU22620]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/> | ||
| + | |||
| + | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[biosynthesis/ acquisition of amino acids]]}} | ||
| − | + | = This gene is a member of the following [[regulons]] = | |
| + | {{SubtiWiki regulon|[[TRAP regulon]]}} | ||
=The gene= | =The gene= | ||
| Line 36: | Line 50: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU22620 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU22620&redirect=T BSU22620] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/trpEDCFBA-hisC-tyrA-aroE.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/trpEDCFBA-hisC-tyrA-aroE.html] | ||
| Line 47: | Line 62: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
=The protein= | =The protein= | ||
| Line 71: | Line 85: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU22620&redirect=T BSU22620] | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1UU0 1UU0] (the enzyme from ''E. coli'') {{PubMed|11518529}} |
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P17731 P17731] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P17731 P17731] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU22620] |
* '''E.C. number:''' [http://www.expasy.org/enzyme/2.6.1.9 2.6.1.9] | * '''E.C. number:''' [http://www.expasy.org/enzyme/2.6.1.9 2.6.1.9] | ||
| Line 88: | Line 103: | ||
=Expression and regulation= | =Expression and regulation= | ||
| + | * '''Operon:''' | ||
| + | ** ''[[aroF]]-[[aroB]]-[[aroH]]-[[trpE]]-[[trpD]]-[[trpC]]-[[trpF]]-[[trpB]]-[[trpA]]-[[hisC]]-[[tyrA]]-[[aroE]]'' (according to [http://dbtbs.hgc.jp/COG/prom/trpEDCFBA-hisC-tyrA-aroE.html DBTBS]) | ||
| + | ** ''[[trpE]]-[[trpD]]-[[trpC]]-[[trpF]]-[[trpB]]-[[trpA]]-[[hisC]]-[[tyrA]]-[[aroE]]'' {{PubMed|3924737}} | ||
| + | ** ''[[hisC]]-[[tyrA]]-[[aroE]]'' (according to [http://dbtbs.hgc.jp/COG/prom/trpEDCFBA-hisC-tyrA-aroE.html DBTBS]) | ||
| − | * ''' | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=hisC_2370415_2371497_-1 hisC] {{PubMed|22383849}} |
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
| + | ** ''[[trpE]]'': [[SigA]] {{PubMed|6436812}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** induction by tryptophan ([[MtrB|TRAP]]) {{PubMed|1551827}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[MtrB|TRAP]]: binding to the mRNA in the presence of tryptophan, this results in transcription termination {{PubMed|8419914}} | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
| + | ** the mRNA is substantially stabilized upon depletion of [[Rny|RNase Y]] {{PubMed|21815947}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 2715 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 2096 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 4023 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 2507 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 6242 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
| Line 118: | Line 145: | ||
=References= | =References= | ||
| + | <pubmed> 4431, 824269 11518529 3924737 6436812 1551827 8419914 21815947 23540922 </pubmed> | ||
| − | + | [[Category:Protein-coding genes]] | |
Latest revision as of 14:08, 17 April 2014
- Description: histidinol-phosphate aminotransferase / tyrosine and phenylalanine aminotransferase
| Gene name | hisC |
| Synonyms | aroJ |
| Essential | no |
| Product | histidinol-phosphate aminotransferase / tyrosine and phenylalanine aminotransferase |
| Function | biosynthesis of aromatic amino acids |
| Gene expression levels in SubtiExpress: hisC | |
| Metabolic function and regulation of this protein in SubtiPathways: hisC | |
| MW, pI | 39 kDa, 5.005 |
| Gene length, protein length | 1080 bp, 360 aa |
| Immediate neighbours | tyrA, trpA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 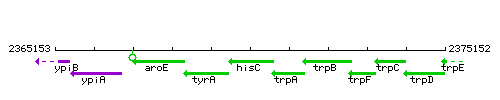
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22620
Phenotypes of a mutant
Database entries
- BsubCyc: BSU22620
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-histidinol phosphate + 2-oxoglutarate = 3-(imidazol-4-yl)-2-oxopropyl phosphate + L-glutamate (according to Swiss-Prot)
- Protein family: bacterial solute-binding protein 3 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU22620
- Swiss prot entry: P17731
- KEGG entry: [3]
- E.C. number: 2.6.1.9
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
- the mRNA is substantially stabilized upon depletion of RNase Y PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 2715 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 2096 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 4023 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 2507 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 6242 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Martin Bommer, John M Ward
A 1-step microplate method for assessing the substrate range of l-α-amino acid aminotransferase.
Enzyme Microb Technol: 2013, 52(4-5);218-25
[PubMed:23540922]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
J Sivaraman, Y Li, R Larocque, J D Schrag, M Cygler, A Matte
Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J Mol Biol: 2001, 311(4);761-76
[PubMed:11518529]
[WorldCat.org]
[DOI]
(P p)
J Otridge, P Gollnick
MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner.
Proc Natl Acad Sci U S A: 1993, 90(1);128-32
[PubMed:8419914]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, P Gollnick, C Yanofsky
The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of tryptophan biosynthesis.
J Bacteriol: 1992, 174(7);2059-64
[PubMed:1551827]
[WorldCat.org]
[DOI]
(P p)
D J Henner, L Band, H Shimotsu
Nucleotide sequence of the Bacillus subtilis tryptophan operon.
Gene: 1985, 34(2-3);169-77
[PubMed:3924737]
[WorldCat.org]
[DOI]
(P p)
H Shimotsu, D J Henner
Characterization of the Bacillus subtilis tryptophan promoter region.
Proc Natl Acad Sci U S A: 1984, 81(20);6315-9
[PubMed:6436812]
[WorldCat.org]
[DOI]
(P p)
D A Weigent, E W Nester
Regulation of histidinol phosphate aminotransferase synthesis by tryptophan in Bacillus subtilis.
J Bacteriol: 1976, 128(1);202-11
[PubMed:824269]
[WorldCat.org]
[DOI]
(P p)
E W Nester, A L Montoya
An enzyme common to histidine and aromatic amino acid biosynthesis in Bacillus subtilis.
J Bacteriol: 1976, 126(2);699-705
[PubMed:4431]
[WorldCat.org]
[DOI]
(P p)