Difference between revisions of "HemA"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || porphyrin biosynthesis | |style="background:#ABCDEF;" align="center"|'''Function''' || porphyrin biosynthesis | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU28170 hemA] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 50 kDa, 5.313 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 50 kDa, 5.313 | ||
Revision as of 14:33, 7 August 2012
- Description: glutamyl-tRNA reductase
| Gene name | hemA |
| Synonyms | |
| Essential | no |
| Product | glutamyl-tRNA reductase |
| Function | porphyrin biosynthesis |
| Gene expression levels in SubtiExpress: hemA | |
| MW, pI | 50 kDa, 5.313 |
| Gene length, protein length | 1365 bp, 455 aa |
| Immediate neighbours | hemX, ysxD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 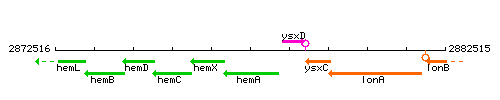
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis of cofactors, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28170
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-glutamate 1-semialdehyde + NADP+ + tRNA(Glu) = L-glutamyl-tRNA(Glu) + NADPH (according to Swiss-Prot)
- Protein family: glutamyl-tRNA reductase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P16618
- KEGG entry: [3]
- E.C. number: 1.2.1.70
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
A F Herbig, J D Helmann
Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA.
Mol Microbiol: 2001, 41(4);849-59
[PubMed:11532148]
[WorldCat.org]
[DOI]
(P p)
Per Johansson, Lars Hederstedt
Organization of genes for tetrapyrrole biosynthesis in gram--positive bacteria.
Microbiology (Reading): 1999, 145 ( Pt 3);529-538
[PubMed:10217486]
[WorldCat.org]
[DOI]
(P p)
N Bsat, L Chen, J D Helmann
Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes.
J Bacteriol: 1996, 178(22);6579-86
[PubMed:8932315]
[WorldCat.org]
[DOI]
(P p)
L Chen, L Keramati, J D Helmann
Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions.
Proc Natl Acad Sci U S A: 1995, 92(18);8190-4
[PubMed:7667267]
[WorldCat.org]
[DOI]
(P p)
I Schröder, P Johansson, L Rutberg, L Hederstedt
The hemX gene of the Bacillus subtilis hemAXCDBL operon encodes a membrane protein, negatively affecting the steady-state cellular concentration of HemA (glutamyl-tRNA reductase).
Microbiology (Reading): 1994, 140 ( Pt 4);731-40
[PubMed:8012594]
[WorldCat.org]
[DOI]
(P p)
I Schröder, L Hederstedt, C G Kannangara, P Gough
Glutamyl-tRNA reductase activity in Bacillus subtilis is dependent on the hemA gene product.
Biochem J: 1992, 281 ( Pt 3)(Pt 3);843-50
[PubMed:1536660]
[WorldCat.org]
[DOI]
(P p)
M Hansson, L Rutberg, I Schröder, L Hederstedt
The Bacillus subtilis hemAXCDBL gene cluster, which encodes enzymes of the biosynthetic pathway from glutamate to uroporphyrinogen III.
J Bacteriol: 1991, 173(8);2590-9
[PubMed:1672867]
[WorldCat.org]
[DOI]
(P p)