Difference between revisions of "Hag"
(→References) |
|||
| Line 59: | Line 59: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | + | * no swarming motility on B medium. [http://www.ncbi.nlm.nih.gov/sites/entrez/19202088 PubMed] | |
| + | * not essential for pellicle biofilm formation, but mutant is outcompeted by the wild-type strain when competed during pellicle formation {{PubMed|26122431}} | ||
| + | * few hours delay in pellicle development {{PubMed|26122431}} | ||
| + | * mutant has increased fitness in planktonic culture when competed with the wild-type NCIB3610 strain {{PubMed|26122431}} | ||
=== Database entries === | === Database entries === | ||
| Line 145: | Line 148: | ||
** 1A783 ( ''hag''::''erm''), {{PubMed|2174860}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A783&Search=1A783 BGSC] | ** 1A783 ( ''hag''::''erm''), {{PubMed|2174860}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A783&Search=1A783 BGSC] | ||
** 1A842 ( ''hag''::''kan''), {{PubMed|14762011}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A842&Search=1A842 BGSC] | ** 1A842 ( ''hag''::''kan''), {{PubMed|14762011}}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A842&Search=1A842 BGSC] | ||
| + | ** DS1677 (marker-less in NCIB3610) {{PubMed|18566286}} | ||
| + | ** TB191 ''amyE''::Phy-''sfgfp'' (marker-less in NCIB3610 with constitutive expressed ''sfgfp'') {{PubMed|26122431}} | ||
| + | ** TB207 ''amyE''::Phy-''mKATE2'' (marker-less in NCIB3610 with constitutive expressed ''mKATE2'') {{PubMed|26122431}} | ||
| + | |||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 158: | Line 165: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | * [[Daniel Kearns]] | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 165: | Line 173: | ||
<pubmed>22672726 25251856 </pubmed> | <pubmed>22672726 25251856 </pubmed> | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed> 23144244 21602220 21856853 2498284,9648743,9096206,7708689,19202088, 18763711, 21895793 10809682 19118355 17555441 20534509 15378759</pubmed> | + | <pubmed> 18566286,26122431,23144244 21602220 21856853 2498284,9648743,9096206,7708689,19202088, 18763711, 21895793 10809682 19118355 17555441 20534509 15378759</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:08, 2 July 2015
- Description: flagellin protein, about 20,000 subunits make up one flagellum
| Gene name | hag |
| Synonyms | |
| Essential | no |
| Product | flagellin protein |
| Function | motility and chemotaxis |
| Gene expression levels in SubtiExpress: hag | |
| Interactions involving this protein in SubtInteract: Hag | |
| Metabolic function and regulation of this protein in SubtiPathways: Hag | |
| MW, pI | 32 kDa, 4.782 |
| Gene length, protein length | 912 bp, 304 aa |
| Immediate neighbours | yvyC, csrA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 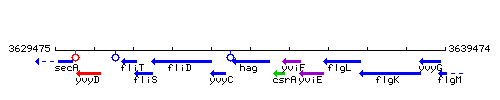
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
motility and chemotaxis, most abundant proteins
This gene is a member of the following regulons
CodY regulon, CsrA regulon, ScoC regulon, SigD regulon
The gene
Basic information
- Locus tag: BSU35360
Phenotypes of a mutant
- no swarming motility on B medium. PubMed
- not essential for pellicle biofilm formation, but mutant is outcompeted by the wild-type strain when competed during pellicle formation PubMed
- few hours delay in pellicle development PubMed
- mutant has increased fitness in planktonic culture when competed with the wild-type NCIB3610 strain PubMed
Database entries
- BsubCyc: BSU35360
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: bacterial flagellin family (according to Swiss-Prot)
- Paralogous protein(s): YvzB (C-terminal domain of Hag)
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU35360
- Structure: 3A5X (from Salmonella typhimurium, 42% identity)
- UniProt: P02968
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
- the hag gene is strongly overexpressed in a ymdB mutant (loss of bistable gene expression) PubMed
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 150241 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 280953 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 9552 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 11147 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 24114 PubMed
Biological materials
- Mutant: GP901 (aphA3), GP902 (tet) PubMed, both available in Jörg Stülke's lab
- 1A915 ( hag::cat), PubMed, available at BGSC
- 1A783 ( hag::erm), PubMed, available at BGSC
- 1A842 ( hag::kan), PubMed, available at BGSC
- DS1677 (marker-less in NCIB3610) PubMed
- TB191 amyE::Phy-sfgfp (marker-less in NCIB3610 with constitutive expressed sfgfp) PubMed
- TB207 amyE::Phy-mKATE2 (marker-less in NCIB3610 with constitutive expressed mKATE2) PubMed
- Expression vector:
- expression in B. subtilis, in pBQ200: pGP1089, available in Jörg Stülke's lab
- lacZ fusion: pGP1035 (in pAC6), pGP755 (in pAC7), there is also a series of promoter deletion variants in pAC6 and pAC7 PubMed, all available in Jörg Stülke's lab
- GFP fusion: BP494 (bglS:: (hag-promoter-cfp-aphA3)), BP496 (amyE:: (hag-promoter-iyfp-cat)), available in Jörg Stülke's lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Sampriti Mukherjee, Daniel B Kearns
The structure and regulation of flagella in Bacillus subtilis.
Annu Rev Genet: 2014, 48;319-40
[PubMed:25251856]
[WorldCat.org]
[DOI]
(I p)
Tony Romeo, Christopher A Vakulskas, Paul Babitzke
Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems.
Environ Microbiol: 2013, 15(2);313-24
[PubMed:22672726]
[WorldCat.org]
[DOI]
(I p)
Original Publications